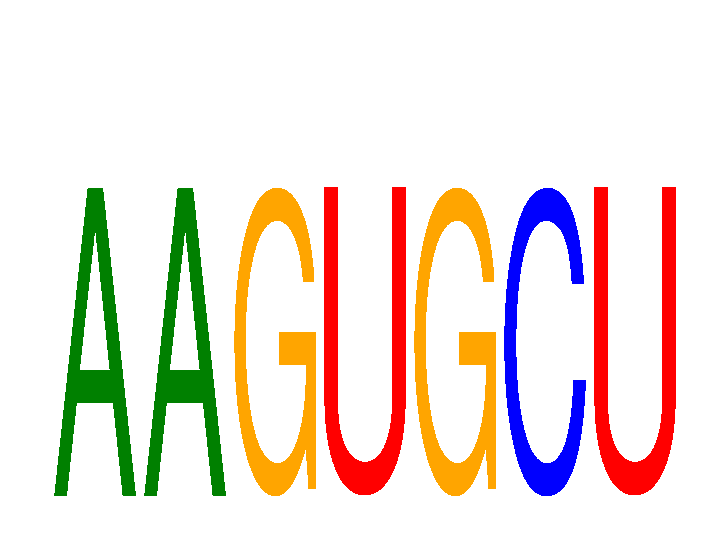Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for AAGUGCU
Z-value: 0.37
miRNA associated with seed AAGUGCU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-302a-3p
|
MIMAT0000684 |
|
hsa-miR-302b-3p
|
MIMAT0000715 |
|
hsa-miR-302c-3p.1
|
MIMAT0000717 |
|
hsa-miR-302d-3p
|
MIMAT0000718 |
|
hsa-miR-302e
|
MIMAT0005931 |
|
hsa-miR-372-3p
|
MIMAT0000724 |
|
hsa-miR-373-3p
|
MIMAT0000726 |
|
hsa-miR-520a-3p
|
MIMAT0002834 |
|
hsa-miR-520b
|
MIMAT0002843 |
|
hsa-miR-520c-3p
|
MIMAT0002846 |
|
hsa-miR-520d-3p
|
MIMAT0002856 |
|
hsa-miR-520e
|
MIMAT0002825 |
Activity profile of AAGUGCU motif
Sorted Z-values of AAGUGCU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGUGCU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_4273751 | 0.61 |
ENST00000675880.1
ENST00000261254.8 |
CCND2
|
cyclin D2 |
| chr4_+_155667198 | 0.40 |
ENST00000296518.11
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr2_+_173354820 | 0.36 |
ENST00000347703.7
ENST00000410101.7 ENST00000410019.3 ENST00000306721.8 |
CDCA7
|
cell division cycle associated 7 |
| chr9_-_96418334 | 0.35 |
ENST00000375256.5
|
ZNF367
|
zinc finger protein 367 |
| chr7_-_41703062 | 0.33 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr4_-_170003738 | 0.33 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr10_+_101131284 | 0.30 |
ENST00000370196.11
ENST00000467928.2 |
TLX1
|
T cell leukemia homeobox 1 |
| chr21_-_31558977 | 0.27 |
ENST00000286827.7
ENST00000541036.5 |
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr3_-_123884290 | 0.24 |
ENST00000346322.9
ENST00000360772.7 ENST00000360304.8 |
MYLK
|
myosin light chain kinase |
| chr6_-_108074703 | 0.24 |
ENST00000193322.8
|
OSTM1
|
osteoclastogenesis associated transmembrane protein 1 |
| chr3_+_47282930 | 0.23 |
ENST00000232766.6
ENST00000437353.5 |
KLHL18
|
kelch like family member 18 |
| chr2_-_227164194 | 0.21 |
ENST00000396625.5
|
COL4A4
|
collagen type IV alpha 4 chain |
| chr1_-_53328053 | 0.21 |
ENST00000371454.6
ENST00000667377.1 ENST00000306052.12 ENST00000668448.1 |
LRP8
|
LDL receptor related protein 8 |
| chr1_-_23531206 | 0.21 |
ENST00000361729.3
|
E2F2
|
E2F transcription factor 2 |
| chr6_+_157381133 | 0.20 |
ENST00000414563.6
ENST00000359775.10 |
ZDHHC14
|
zinc finger DHHC-type palmitoyltransferase 14 |
| chr8_-_123396412 | 0.20 |
ENST00000287394.10
|
ATAD2
|
ATPase family AAA domain containing 2 |
| chr12_-_24949026 | 0.20 |
ENST00000539780.5
ENST00000546285.1 ENST00000342945.9 ENST00000261192.12 |
BCAT1
|
branched chain amino acid transaminase 1 |
| chr4_-_110198650 | 0.20 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr3_-_121545962 | 0.20 |
ENST00000264233.6
|
POLQ
|
DNA polymerase theta |
| chr16_-_71724700 | 0.19 |
ENST00000568954.5
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr12_-_56258327 | 0.18 |
ENST00000267116.8
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr3_-_48188356 | 0.18 |
ENST00000351231.7
ENST00000437972.1 ENST00000302506.8 |
CDC25A
|
cell division cycle 25A |
| chr6_-_31902041 | 0.18 |
ENST00000375527.3
|
ZBTB12
|
zinc finger and BTB domain containing 12 |
| chr11_-_64972070 | 0.18 |
ENST00000301896.6
|
MAJIN
|
membrane anchored junction protein |
| chr10_+_52314272 | 0.17 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr12_+_32502114 | 0.16 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_+_30606574 | 0.16 |
ENST00000295754.10
ENST00000359013.4 |
TGFBR2
|
transforming growth factor beta receptor 2 |
| chr1_+_107056656 | 0.15 |
ENST00000370078.2
|
PRMT6
|
protein arginine methyltransferase 6 |
| chrX_-_20266834 | 0.15 |
ENST00000379565.9
|
RPS6KA3
|
ribosomal protein S6 kinase A3 |
| chr3_-_171460368 | 0.15 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr9_-_16870662 | 0.15 |
ENST00000380672.9
|
BNC2
|
basonuclin 2 |
| chr3_-_197749688 | 0.15 |
ENST00000273582.9
|
RUBCN
|
rubicon autophagy regulator |
| chr3_-_165196369 | 0.15 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr20_+_58309704 | 0.15 |
ENST00000244040.4
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr17_-_45490696 | 0.15 |
ENST00000430334.8
ENST00000584420.1 ENST00000589780.5 |
PLEKHM1
|
pleckstrin homology and RUN domain containing M1 |
| chr11_+_69641146 | 0.14 |
ENST00000227507.3
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr3_+_58306236 | 0.14 |
ENST00000295959.10
ENST00000445193.7 ENST00000466547.1 ENST00000475412.5 ENST00000474660.5 ENST00000477305.5 ENST00000481972.5 |
RPP14
HTD2
|
ribonuclease P/MRP subunit p14 hydroxyacyl-thioester dehydratase type 2 |
| chr4_-_162163989 | 0.14 |
ENST00000306100.10
ENST00000427802.2 |
FSTL5
|
follistatin like 5 |
| chr19_-_14136553 | 0.14 |
ENST00000592798.5
ENST00000474890.1 ENST00000263382.8 |
ASF1B
|
anti-silencing function 1B histone chaperone |
| chr4_+_44678412 | 0.14 |
ENST00000281543.6
|
GUF1
|
GTP binding elongation factor GUF1 |
| chrX_+_16719595 | 0.14 |
ENST00000380155.4
|
SYAP1
|
synapse associated protein 1 |
| chr2_+_99337364 | 0.13 |
ENST00000617677.1
ENST00000289371.11 |
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr15_-_61229297 | 0.13 |
ENST00000335670.11
|
RORA
|
RAR related orphan receptor A |
| chr3_+_126983035 | 0.13 |
ENST00000393409.3
|
PLXNA1
|
plexin A1 |
| chr5_-_115544734 | 0.13 |
ENST00000274457.5
|
FEM1C
|
fem-1 homolog C |
| chr21_-_43427131 | 0.13 |
ENST00000270162.8
|
SIK1
|
salt inducible kinase 1 |
| chrX_+_28587411 | 0.13 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chrX_+_24149629 | 0.13 |
ENST00000428571.5
ENST00000539115.5 |
ZFX
|
zinc finger protein X-linked |
| chr6_-_105137147 | 0.13 |
ENST00000314641.10
|
BVES
|
blood vessel epicardial substance |
| chr9_-_131270493 | 0.12 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr2_-_25878445 | 0.12 |
ENST00000336112.9
ENST00000435504.9 |
ASXL2
|
ASXL transcriptional regulator 2 |
| chr9_-_122227525 | 0.12 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr11_+_129375841 | 0.12 |
ENST00000281437.6
|
BARX2
|
BARX homeobox 2 |
| chr3_+_119294337 | 0.12 |
ENST00000264245.9
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr18_+_23689439 | 0.12 |
ENST00000313654.14
|
LAMA3
|
laminin subunit alpha 3 |
| chr1_-_179229671 | 0.12 |
ENST00000502732.6
ENST00000392043.4 |
ABL2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr10_-_121927979 | 0.11 |
ENST00000369040.8
ENST00000369043.8 ENST00000224652.11 |
ATE1
|
arginyltransferase 1 |
| chr17_-_68291116 | 0.11 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chr20_+_1894145 | 0.11 |
ENST00000400068.7
|
SIRPA
|
signal regulatory protein alpha |
| chr8_+_105318428 | 0.11 |
ENST00000407775.7
|
ZFPM2
|
zinc finger protein, FOG family member 2 |
| chr4_+_73740541 | 0.11 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr7_+_28412511 | 0.11 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr15_-_83284645 | 0.11 |
ENST00000345382.7
|
BNC1
|
basonuclin 1 |
| chr10_-_79445617 | 0.11 |
ENST00000372336.4
|
ZCCHC24
|
zinc finger CCHC-type containing 24 |
| chr15_+_31326807 | 0.11 |
ENST00000307145.4
|
KLF13
|
Kruppel like factor 13 |
| chr1_-_20486197 | 0.10 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr2_-_131093378 | 0.10 |
ENST00000409185.5
ENST00000389915.4 |
FAM168B
|
family with sequence similarity 168 member B |
| chr16_+_29973847 | 0.10 |
ENST00000308893.9
|
TAOK2
|
TAO kinase 2 |
| chr1_+_3069160 | 0.10 |
ENST00000511072.5
|
PRDM16
|
PR/SET domain 16 |
| chr17_+_68035722 | 0.10 |
ENST00000679078.1
ENST00000330459.8 ENST00000584026.6 |
KPNA2
|
karyopherin subunit alpha 2 |
| chr15_+_92393841 | 0.10 |
ENST00000268164.8
ENST00000539113.5 ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr2_+_134120169 | 0.10 |
ENST00000409645.5
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase |
| chr11_+_122655712 | 0.10 |
ENST00000284273.6
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr2_+_149330506 | 0.10 |
ENST00000334166.9
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr19_+_16197900 | 0.09 |
ENST00000429941.6
ENST00000291439.8 ENST00000444449.6 ENST00000589822.5 |
AP1M1
|
adaptor related protein complex 1 subunit mu 1 |
| chr5_+_98769273 | 0.09 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr8_-_130443581 | 0.09 |
ENST00000357668.2
ENST00000518721.6 |
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr2_+_69915100 | 0.09 |
ENST00000264444.7
|
MXD1
|
MAX dimerization protein 1 |
| chr5_+_179820895 | 0.09 |
ENST00000504627.1
ENST00000389805.9 ENST00000510187.5 |
SQSTM1
|
sequestosome 1 |
| chr5_+_72107453 | 0.09 |
ENST00000296755.12
ENST00000511641.2 |
MAP1B
|
microtubule associated protein 1B |
| chr7_-_20217342 | 0.09 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr1_-_227947924 | 0.09 |
ENST00000272164.6
|
WNT9A
|
Wnt family member 9A |
| chr4_-_41214602 | 0.09 |
ENST00000508676.5
ENST00000506352.5 ENST00000295974.12 |
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr2_+_26848093 | 0.09 |
ENST00000288699.11
|
DPYSL5
|
dihydropyrimidinase like 5 |
| chr14_+_93430853 | 0.09 |
ENST00000553484.5
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr7_+_6104881 | 0.09 |
ENST00000306177.9
ENST00000465073.6 |
USP42
|
ubiquitin specific peptidase 42 |
| chr6_+_41072939 | 0.08 |
ENST00000341376.11
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y subunit alpha |
| chr8_+_1823967 | 0.08 |
ENST00000520359.5
ENST00000518288.5 |
ARHGEF10
|
Rho guanine nucleotide exchange factor 10 |
| chr10_+_96043394 | 0.08 |
ENST00000403870.7
ENST00000265992.9 ENST00000465148.3 |
CCNJ
|
cyclin J |
| chr6_+_157823191 | 0.08 |
ENST00000681534.1
ENST00000681183.1 ENST00000679732.1 ENST00000681186.1 ENST00000680078.1 ENST00000681138.1 ENST00000680495.1 ENST00000392185.8 |
SNX9
|
sorting nexin 9 |
| chr20_-_9838831 | 0.08 |
ENST00000378423.5
|
PAK5
|
p21 (RAC1) activated kinase 5 |
| chr9_+_74497308 | 0.08 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr9_-_19102887 | 0.08 |
ENST00000380502.8
|
HAUS6
|
HAUS augmin like complex subunit 6 |
| chr17_+_75721451 | 0.08 |
ENST00000200181.8
|
ITGB4
|
integrin subunit beta 4 |
| chr14_+_70641896 | 0.08 |
ENST00000256367.3
|
TTC9
|
tetratricopeptide repeat domain 9 |
| chr11_-_78341876 | 0.08 |
ENST00000340149.6
|
GAB2
|
GRB2 associated binding protein 2 |
| chr6_+_20401864 | 0.08 |
ENST00000346618.8
ENST00000613242.4 |
E2F3
|
E2F transcription factor 3 |
| chr5_-_168579319 | 0.07 |
ENST00000522176.1
ENST00000239231.7 |
PANK3
|
pantothenate kinase 3 |
| chr12_-_107761113 | 0.07 |
ENST00000228437.10
|
PRDM4
|
PR/SET domain 4 |
| chr14_-_99272184 | 0.07 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr2_+_197804583 | 0.07 |
ENST00000428675.6
|
PLCL1
|
phospholipase C like 1 (inactive) |
| chr11_+_61752603 | 0.07 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr22_+_21417357 | 0.07 |
ENST00000407464.7
|
HIC2
|
HIC ZBTB transcriptional repressor 2 |
| chr20_-_32207708 | 0.07 |
ENST00000246229.5
|
PLAGL2
|
PLAG1 like zinc finger 2 |
| chr11_+_9384621 | 0.07 |
ENST00000379719.8
ENST00000527431.1 ENST00000630083.1 |
IPO7
|
importin 7 |
| chr3_+_196739839 | 0.07 |
ENST00000327134.7
|
PAK2
|
p21 (RAC1) activated kinase 2 |
| chr11_+_33257265 | 0.07 |
ENST00000303296.9
ENST00000379016.7 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr12_+_56521798 | 0.07 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr1_-_115841116 | 0.07 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2 |
| chrX_+_77447387 | 0.07 |
ENST00000439435.3
|
FGF16
|
fibroblast growth factor 16 |
| chr9_-_23821275 | 0.07 |
ENST00000380110.8
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr19_-_4066892 | 0.07 |
ENST00000322357.9
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr17_+_28335718 | 0.07 |
ENST00000226225.7
|
TNFAIP1
|
TNF alpha induced protein 1 |
| chr12_+_111034136 | 0.07 |
ENST00000261726.11
|
CUX2
|
cut like homeobox 2 |
| chr19_+_4402615 | 0.07 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr14_+_52552830 | 0.07 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr5_-_115180037 | 0.07 |
ENST00000514154.1
ENST00000282369.7 |
TRIM36
|
tripartite motif containing 36 |
| chr12_+_55966821 | 0.07 |
ENST00000553376.5
ENST00000440311.6 ENST00000266970.9 ENST00000354056.4 |
CDK2
|
cyclin dependent kinase 2 |
| chr17_-_42154916 | 0.07 |
ENST00000592574.1
ENST00000550406.1 ENST00000547517.5 ENST00000346213.9 ENST00000393860.7 |
ENSG00000267261.5
RAB5C
|
novel protein RAB5C, member RAS oncogene family |
| chr14_+_32939243 | 0.07 |
ENST00000346562.6
ENST00000548645.5 ENST00000356141.8 ENST00000357798.9 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr12_+_71754834 | 0.07 |
ENST00000261263.5
|
RAB21
|
RAB21, member RAS oncogene family |
| chr16_-_4273014 | 0.07 |
ENST00000204517.11
|
TFAP4
|
transcription factor AP-4 |
| chr20_+_35226676 | 0.07 |
ENST00000246186.8
|
MMP24
|
matrix metallopeptidase 24 |
| chr12_-_21774688 | 0.07 |
ENST00000240662.3
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr9_+_36036899 | 0.06 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr10_+_28677487 | 0.06 |
ENST00000375533.6
|
BAMBI
|
BMP and activin membrane bound inhibitor |
| chr4_-_78939352 | 0.06 |
ENST00000512733.5
|
PAQR3
|
progestin and adipoQ receptor family member 3 |
| chr1_-_55215345 | 0.06 |
ENST00000294383.7
|
USP24
|
ubiquitin specific peptidase 24 |
| chr2_+_102619531 | 0.06 |
ENST00000233969.3
|
SLC9A2
|
solute carrier family 9 member A2 |
| chr12_+_55019967 | 0.06 |
ENST00000242994.4
|
NEUROD4
|
neuronal differentiation 4 |
| chr3_+_14947568 | 0.06 |
ENST00000413118.5
ENST00000425241.5 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr10_+_14878848 | 0.06 |
ENST00000433779.5
ENST00000378325.7 ENST00000354919.11 ENST00000313519.9 ENST00000420416.1 |
SUV39H2
|
suppressor of variegation 3-9 homolog 2 |
| chr20_-_37095985 | 0.06 |
ENST00000344359.7
ENST00000373664.8 |
RBL1
|
RB transcriptional corepressor like 1 |
| chr22_-_50474942 | 0.06 |
ENST00000348911.10
ENST00000380817.8 |
SBF1
|
SET binding factor 1 |
| chr6_-_166627244 | 0.06 |
ENST00000265678.9
|
RPS6KA2
|
ribosomal protein S6 kinase A2 |
| chr6_+_11537738 | 0.06 |
ENST00000379426.2
|
TMEM170B
|
transmembrane protein 170B |
| chr17_-_55722857 | 0.06 |
ENST00000424486.3
|
TMEM100
|
transmembrane protein 100 |
| chr4_+_127782270 | 0.06 |
ENST00000508549.5
ENST00000296464.9 |
HSPA4L
|
heat shock protein family A (Hsp70) member 4 like |
| chr20_-_675793 | 0.06 |
ENST00000488788.2
ENST00000246104.7 |
ENSG00000270299.1
SCRT2
|
novel protein scratch family transcriptional repressor 2 |
| chr7_+_129434424 | 0.06 |
ENST00000249344.7
ENST00000435494.2 |
STRIP2
|
striatin interacting protein 2 |
| chr20_-_43189733 | 0.06 |
ENST00000373187.5
ENST00000356100.6 ENST00000373184.5 ENST00000373190.5 |
PTPRT
|
protein tyrosine phosphatase receptor type T |
| chr5_+_90474879 | 0.06 |
ENST00000504930.5
ENST00000514483.5 |
POLR3G
|
RNA polymerase III subunit G |
| chr2_-_43226594 | 0.06 |
ENST00000282388.4
|
ZFP36L2
|
ZFP36 ring finger protein like 2 |
| chrX_-_25015924 | 0.06 |
ENST00000379044.5
|
ARX
|
aristaless related homeobox |
| chr3_-_50567646 | 0.06 |
ENST00000426034.5
ENST00000441239.5 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr2_-_11344580 | 0.06 |
ENST00000616279.4
ENST00000315872.11 |
ROCK2
|
Rho associated coiled-coil containing protein kinase 2 |
| chr14_-_53152371 | 0.06 |
ENST00000323669.10
|
DDHD1
|
DDHD domain containing 1 |
| chr17_+_4833331 | 0.06 |
ENST00000355280.11
ENST00000347992.11 |
MINK1
|
misshapen like kinase 1 |
| chr1_-_119648165 | 0.06 |
ENST00000421812.3
|
ZNF697
|
zinc finger protein 697 |
| chr16_-_2135898 | 0.06 |
ENST00000262304.9
ENST00000423118.5 |
PKD1
|
polycystin 1, transient receptor potential channel interacting |
| chr2_+_73984902 | 0.06 |
ENST00000409262.8
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr2_+_169827432 | 0.06 |
ENST00000272793.11
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr7_+_120273129 | 0.06 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
| chr2_-_171894227 | 0.06 |
ENST00000422440.7
|
SLC25A12
|
solute carrier family 25 member 12 |
| chr4_+_159267737 | 0.05 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr12_-_16608183 | 0.05 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr1_-_84690406 | 0.05 |
ENST00000605755.5
ENST00000342203.8 ENST00000437941.6 |
SSX2IP
|
SSX family member 2 interacting protein |
| chr18_+_57352541 | 0.05 |
ENST00000324000.4
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr1_+_11980181 | 0.05 |
ENST00000444836.5
ENST00000674817.1 ENST00000675053.1 ENST00000675817.1 ENST00000675298.1 ENST00000676369.1 ENST00000412236.2 ENST00000675530.1 ENST00000674548.1 ENST00000674658.1 ENST00000674910.1 ENST00000675231.1 |
MFN2
|
mitofusin 2 |
| chr17_+_46851580 | 0.05 |
ENST00000290015.7
ENST00000393461.2 |
WNT9B
|
Wnt family member 9B |
| chr2_-_69643703 | 0.05 |
ENST00000406297.7
ENST00000409085.9 |
AAK1
|
AP2 associated kinase 1 |
| chr2_-_2331225 | 0.05 |
ENST00000648627.1
ENST00000649663.1 ENST00000650560.1 ENST00000428368.7 ENST00000648316.1 ENST00000648665.1 ENST00000649313.1 ENST00000399161.7 ENST00000647738.2 |
MYT1L
|
myelin transcription factor 1 like |
| chr5_-_142325001 | 0.05 |
ENST00000344120.4
ENST00000434127.3 |
SPRY4
|
sprouty RTK signaling antagonist 4 |
| chr1_-_37859583 | 0.05 |
ENST00000373036.5
|
MTF1
|
metal regulatory transcription factor 1 |
| chr10_+_100535927 | 0.05 |
ENST00000299163.7
|
HIF1AN
|
hypoxia inducible factor 1 subunit alpha inhibitor |
| chr1_-_225889143 | 0.05 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr22_+_29073024 | 0.05 |
ENST00000400335.9
|
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr2_-_234497035 | 0.05 |
ENST00000390645.2
ENST00000339728.6 |
ARL4C
|
ADP ribosylation factor like GTPase 4C |
| chr20_+_44475867 | 0.05 |
ENST00000262605.9
ENST00000372904.7 ENST00000372906.2 ENST00000456317.1 |
TTPAL
|
alpha tocopherol transfer protein like |
| chr2_-_224585354 | 0.05 |
ENST00000264414.9
ENST00000344951.8 |
CUL3
|
cullin 3 |
| chr8_+_42896883 | 0.05 |
ENST00000307602.9
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr21_-_34888683 | 0.05 |
ENST00000344691.8
ENST00000358356.9 |
RUNX1
|
RUNX family transcription factor 1 |
| chr7_-_28180735 | 0.05 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr2_-_20225123 | 0.05 |
ENST00000254351.9
|
SDC1
|
syndecan 1 |
| chr7_-_13989658 | 0.05 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chr3_-_56801939 | 0.05 |
ENST00000296315.8
ENST00000495373.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr8_-_90646074 | 0.05 |
ENST00000458549.7
|
TMEM64
|
transmembrane protein 64 |
| chr8_-_37899454 | 0.05 |
ENST00000522727.5
ENST00000287263.8 ENST00000330843.9 |
RAB11FIP1
|
RAB11 family interacting protein 1 |
| chr3_+_46979659 | 0.05 |
ENST00000450053.8
|
NBEAL2
|
neurobeachin like 2 |
| chr8_+_94641145 | 0.05 |
ENST00000433389.8
ENST00000358397.9 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr10_-_110304894 | 0.05 |
ENST00000369603.10
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr13_-_26760741 | 0.05 |
ENST00000405846.5
|
GPR12
|
G protein-coupled receptor 12 |
| chr8_-_100952918 | 0.05 |
ENST00000395957.6
ENST00000395948.6 ENST00000457309.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr3_-_12967668 | 0.05 |
ENST00000273221.8
|
IQSEC1
|
IQ motif and Sec7 domain ArfGEF 1 |
| chr20_+_34704336 | 0.05 |
ENST00000374809.6
ENST00000374810.8 ENST00000451665.5 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr1_+_213987929 | 0.05 |
ENST00000498508.6
ENST00000366958.9 |
PROX1
|
prospero homeobox 1 |
| chr10_-_118754956 | 0.04 |
ENST00000369151.8
|
CACUL1
|
CDK2 associated cullin domain 1 |
| chr19_-_17075418 | 0.04 |
ENST00000253669.10
|
HAUS8
|
HAUS augmin like complex subunit 8 |
| chr6_-_52284677 | 0.04 |
ENST00000596288.7
ENST00000616552.4 ENST00000229854.12 ENST00000419835.8 |
MCM3
|
minichromosome maintenance complex component 3 |
| chr5_-_135452318 | 0.04 |
ENST00000537858.2
|
TIFAB
|
TIFA inhibitor |
| chr13_-_25172278 | 0.04 |
ENST00000515384.2
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr1_-_94237562 | 0.04 |
ENST00000260526.11
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr5_+_75511756 | 0.04 |
ENST00000241436.8
|
POLK
|
DNA polymerase kappa |
| chr12_-_44875647 | 0.04 |
ENST00000395487.6
|
NELL2
|
neural EGFL like 2 |
| chr3_+_11272413 | 0.04 |
ENST00000446450.6
ENST00000354956.9 ENST00000354449.7 ENST00000419112.5 |
ATG7
|
autophagy related 7 |
| chr13_-_21061492 | 0.04 |
ENST00000382592.5
|
LATS2
|
large tumor suppressor kinase 2 |
| chr14_-_77498808 | 0.04 |
ENST00000342219.9
ENST00000493585.5 ENST00000554801.2 |
ISM2
|
isthmin 2 |
| chrX_+_14873399 | 0.04 |
ENST00000380492.8
ENST00000482354.5 |
MOSPD2
|
motile sperm domain containing 2 |
| chr3_+_172750682 | 0.04 |
ENST00000232458.9
ENST00000540509.5 |
ECT2
|
epithelial cell transforming 2 |
| chr17_+_7281711 | 0.04 |
ENST00000317370.13
ENST00000571308.5 |
SLC2A4
|
solute carrier family 2 member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.2 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.3 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 0.0 | 0.6 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0071626 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) metanephric proximal tubule development(GO:0072237) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.0 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.0 | GO:0071630 | trophectodermal cellular morphogenesis(GO:0001831) nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.0 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 0.0 | 0.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:2000670 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:1902617 | late nucleophagy(GO:0044805) negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) response to fluoride(GO:1902617) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0061428 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |