Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for AIRE
Z-value: 0.72
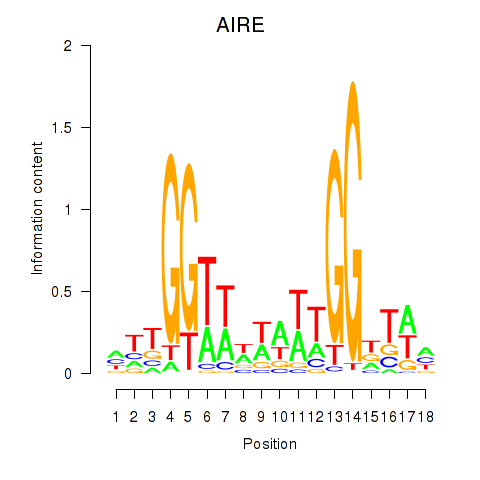
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.17 | AIRE |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg38_v1_chr21_+_44285869_44285884 | 0.27 | 1.4e-01 | Click! |
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_75480800 | 1.48 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr9_+_87497222 | 1.45 |
ENST00000358077.9
|
DAPK1
|
death associated protein kinase 1 |
| chr20_+_59628609 | 1.23 |
ENST00000541461.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr12_+_75481204 | 1.17 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr12_+_75480745 | 1.11 |
ENST00000266659.8
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr17_+_7306975 | 1.08 |
ENST00000336452.11
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr18_+_63775369 | 0.89 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr10_-_100081854 | 0.87 |
ENST00000370418.8
|
CPN1
|
carboxypeptidase N subunit 1 |
| chr19_+_55857437 | 0.81 |
ENST00000587891.5
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chr18_+_63775395 | 0.75 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr9_-_65285209 | 0.74 |
ENST00000377420.1
|
FOXD4L5
|
forkhead box D4 like 5 |
| chr11_-_66907891 | 0.68 |
ENST00000393955.6
|
PC
|
pyruvate carboxylase |
| chr11_+_35189964 | 0.65 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr19_-_23687163 | 0.59 |
ENST00000601010.5
ENST00000601935.5 ENST00000600313.5 ENST00000596211.5 ENST00000359788.9 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr2_-_189179754 | 0.57 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr16_-_20697680 | 0.54 |
ENST00000520010.6
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr8_+_53851786 | 0.54 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr3_-_48089203 | 0.52 |
ENST00000468075.2
ENST00000360240.10 |
MAP4
|
microtubule associated protein 4 |
| chr4_-_119322128 | 0.52 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr6_-_39322968 | 0.52 |
ENST00000507712.5
|
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr12_-_24949026 | 0.52 |
ENST00000539780.5
ENST00000546285.1 ENST00000342945.9 ENST00000261192.12 |
BCAT1
|
branched chain amino acid transaminase 1 |
| chr2_-_237414127 | 0.51 |
ENST00000472056.5
|
COL6A3
|
collagen type VI alpha 3 chain |
| chrM_+_12329 | 0.49 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr8_+_127736046 | 0.46 |
ENST00000641036.1
ENST00000377970.6 |
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr10_-_96359273 | 0.45 |
ENST00000393871.5
ENST00000419479.5 ENST00000393870.3 |
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr4_+_94489030 | 0.45 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
| chr4_-_87529460 | 0.45 |
ENST00000418378.5
|
SPARCL1
|
SPARC like 1 |
| chr16_+_11345429 | 0.45 |
ENST00000576027.1
ENST00000312499.6 ENST00000648619.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr2_-_208129824 | 0.44 |
ENST00000282141.4
|
CRYGC
|
crystallin gamma C |
| chr3_-_121749704 | 0.44 |
ENST00000393667.7
ENST00000340645.9 ENST00000614479.4 |
GOLGB1
|
golgin B1 |
| chr5_-_78549151 | 0.44 |
ENST00000515007.6
|
LHFPL2
|
LHFPL tetraspan subfamily member 2 |
| chr7_-_66995576 | 0.43 |
ENST00000246868.7
ENST00000617799.1 |
SBDS
|
SBDS ribosome maturation factor |
| chr20_+_56248732 | 0.43 |
ENST00000243911.2
|
MC3R
|
melanocortin 3 receptor |
| chr9_-_21482313 | 0.41 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr14_-_81436447 | 0.40 |
ENST00000649389.1
ENST00000557055.5 |
STON2
|
stonin 2 |
| chr2_-_224947030 | 0.40 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chrX_+_154144242 | 0.39 |
ENST00000369951.9
|
OPN1LW
|
opsin 1, long wave sensitive |
| chr7_-_36724457 | 0.39 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr11_-_71821548 | 0.38 |
ENST00000525199.1
|
ZNF705E
|
zinc finger protein 705E |
| chr6_-_137044269 | 0.38 |
ENST00000635289.1
ENST00000541547.5 |
IL20RA
|
interleukin 20 receptor subunit alpha |
| chr14_-_106511856 | 0.38 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr7_-_36724543 | 0.38 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr21_+_29130630 | 0.37 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr5_+_122845488 | 0.37 |
ENST00000513881.5
|
SNX24
|
sorting nexin 24 |
| chr15_-_99249523 | 0.37 |
ENST00000560235.1
ENST00000394132.7 ENST00000560860.5 ENST00000558078.5 ENST00000560772.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr7_-_36724380 | 0.36 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr8_-_7452365 | 0.34 |
ENST00000458665.5
ENST00000528168.3 |
SPAG11B
|
sperm associated antigen 11B |
| chr3_-_139480723 | 0.34 |
ENST00000511956.1
ENST00000506825.1 |
RBP2
|
retinol binding protein 2 |
| chr14_-_100569780 | 0.33 |
ENST00000355173.7
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr12_+_20695553 | 0.33 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr1_+_165895583 | 0.31 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr8_+_103372388 | 0.31 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr21_+_38272291 | 0.31 |
ENST00000438657.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr14_-_64942720 | 0.30 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chr10_+_89283685 | 0.30 |
ENST00000638108.1
|
IFIT2
|
interferon induced protein with tetratricopeptide repeats 2 |
| chr20_+_46008900 | 0.30 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr19_-_10380454 | 0.29 |
ENST00000530829.1
ENST00000529370.5 |
TYK2
|
tyrosine kinase 2 |
| chr15_+_22094522 | 0.29 |
ENST00000328795.5
|
OR4N4
|
olfactory receptor family 4 subfamily N member 4 |
| chr19_+_44914833 | 0.29 |
ENST00000589078.1
ENST00000586638.5 |
APOC1
|
apolipoprotein C1 |
| chr3_+_136819069 | 0.29 |
ENST00000393079.3
ENST00000446465.3 |
SLC35G2
|
solute carrier family 35 member G2 |
| chr6_+_36871841 | 0.29 |
ENST00000359359.6
|
C6orf89
|
chromosome 6 open reading frame 89 |
| chr17_+_3420568 | 0.28 |
ENST00000574571.4
|
OR3A3
|
olfactory receptor family 3 subfamily A member 3 |
| chrM_-_14669 | 0.28 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 |
| chr21_-_6467509 | 0.27 |
ENST00000624406.3
ENST00000398168.5 ENST00000624934.3 |
CBSL
|
cystathionine beta-synthase like |
| chrX_-_49184789 | 0.27 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr4_-_174829212 | 0.27 |
ENST00000340217.5
ENST00000274093.8 |
GLRA3
|
glycine receptor alpha 3 |
| chr1_+_111473972 | 0.26 |
ENST00000369718.4
|
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_+_15153698 | 0.26 |
ENST00000400796.7
ENST00000376008.3 ENST00000434578.6 |
TMEM51
|
transmembrane protein 51 |
| chr17_-_40994159 | 0.26 |
ENST00000391586.3
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr2_-_165204042 | 0.26 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr12_-_64390727 | 0.26 |
ENST00000543942.7
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr13_-_46182136 | 0.25 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr14_-_24442765 | 0.25 |
ENST00000555365.5
ENST00000399395.8 ENST00000553930.5 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U member 1 |
| chr8_-_15238423 | 0.24 |
ENST00000382080.6
|
SGCZ
|
sarcoglycan zeta |
| chr17_+_47522931 | 0.24 |
ENST00000525007.5
ENST00000530173.6 |
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr2_-_165953750 | 0.24 |
ENST00000243344.8
ENST00000679799.1 ENST00000679840.1 ENST00000681606.1 ENST00000680448.1 |
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr19_-_43883964 | 0.24 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr16_+_3204247 | 0.23 |
ENST00000304646.2
|
OR1F1
|
olfactory receptor family 1 subfamily F member 1 |
| chr15_+_21651844 | 0.23 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr6_-_26043704 | 0.23 |
ENST00000615966.2
|
H2BC3
|
H2B clustered histone 3 |
| chr9_-_92536031 | 0.23 |
ENST00000344604.9
ENST00000375540.5 |
ECM2
|
extracellular matrix protein 2 |
| chr19_-_3600581 | 0.22 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr2_+_11534039 | 0.22 |
ENST00000381486.7
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr1_-_16431371 | 0.22 |
ENST00000612240.1
|
SPATA21
|
spermatogenesis associated 21 |
| chr19_-_35228699 | 0.21 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B |
| chr6_-_11044275 | 0.21 |
ENST00000354666.4
|
ELOVL2
|
ELOVL fatty acid elongase 2 |
| chr5_-_64768619 | 0.21 |
ENST00000513458.9
|
SREK1IP1
|
SREK1 interacting protein 1 |
| chr5_+_122845584 | 0.21 |
ENST00000395451.8
ENST00000261369.9 ENST00000506996.5 |
SNX24
|
sorting nexin 24 |
| chr11_+_6926417 | 0.20 |
ENST00000610573.4
ENST00000278319.10 |
ZNF215
|
zinc finger protein 215 |
| chr19_-_58573555 | 0.20 |
ENST00000599369.5
|
MZF1
|
myeloid zinc finger 1 |
| chr2_+_29113989 | 0.19 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr13_+_52455429 | 0.19 |
ENST00000468284.1
ENST00000378034.7 ENST00000378037.9 ENST00000258607.10 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr17_+_79034185 | 0.18 |
ENST00000581774.5
|
C1QTNF1
|
C1q and TNF related 1 |
| chr19_+_44914702 | 0.18 |
ENST00000592885.5
ENST00000589781.1 |
APOC1
|
apolipoprotein C1 |
| chr20_-_35147285 | 0.18 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr19_-_46746421 | 0.18 |
ENST00000263280.11
|
STRN4
|
striatin 4 |
| chr19_-_16496156 | 0.18 |
ENST00000269881.8
|
CALR3
|
calreticulin 3 |
| chr14_-_24442662 | 0.18 |
ENST00000554698.5
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U member 1 |
| chr16_-_31135699 | 0.18 |
ENST00000317508.11
ENST00000568261.5 ENST00000567797.1 |
PRSS8
|
serine protease 8 |
| chr3_-_33645433 | 0.17 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr19_+_44914588 | 0.17 |
ENST00000592535.6
|
APOC1
|
apolipoprotein C1 |
| chr17_-_3398410 | 0.17 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor family 1 subfamily E member 1 |
| chr21_+_38272410 | 0.17 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr1_-_243163310 | 0.17 |
ENST00000492145.1
ENST00000490813.5 ENST00000464936.5 |
CEP170
|
centrosomal protein 170 |
| chr11_-_5324297 | 0.16 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2 |
| chr21_+_38272250 | 0.16 |
ENST00000398932.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr14_+_22105305 | 0.16 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr1_+_207496147 | 0.16 |
ENST00000400960.7
ENST00000367049.9 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr3_-_88149815 | 0.15 |
ENST00000467332.1
ENST00000462901.5 |
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_+_233778330 | 0.15 |
ENST00000389758.3
|
MROH2A
|
maestro heat like repeat family member 2A |
| chr2_-_135876382 | 0.15 |
ENST00000264156.3
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr17_-_40565459 | 0.15 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
C-C motif chemokine receptor 7 |
| chr17_+_56153458 | 0.15 |
ENST00000318698.6
ENST00000682825.1 ENST00000566473.6 |
ANKFN1
|
ankyrin repeat and fibronectin type III domain containing 1 |
| chr2_-_240820945 | 0.14 |
ENST00000428768.2
ENST00000650053.1 ENST00000650130.1 |
KIF1A
|
kinesin family member 1A |
| chr1_-_51878799 | 0.14 |
ENST00000354831.11
ENST00000544028.5 |
NRDC
|
nardilysin convertase |
| chr19_+_13151975 | 0.13 |
ENST00000588173.1
|
IER2
|
immediate early response 2 |
| chr2_-_222656067 | 0.13 |
ENST00000281828.8
|
FARSB
|
phenylalanyl-tRNA synthetase subunit beta |
| chr3_-_36739791 | 0.13 |
ENST00000416516.2
|
DCLK3
|
doublecortin like kinase 3 |
| chr12_-_10986912 | 0.13 |
ENST00000506868.1
|
TAS2R50
|
taste 2 receptor member 50 |
| chr8_+_90940517 | 0.12 |
ENST00000521366.1
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1 |
| chr1_+_162497805 | 0.11 |
ENST00000538489.5
ENST00000489294.2 |
UHMK1
|
U2AF homology motif kinase 1 |
| chr1_+_241652275 | 0.11 |
ENST00000366552.6
ENST00000437684.7 |
WDR64
|
WD repeat domain 64 |
| chr22_+_24806265 | 0.11 |
ENST00000400359.4
|
SGSM1
|
small G protein signaling modulator 1 |
| chr11_-_73876674 | 0.11 |
ENST00000545127.1
ENST00000537289.1 ENST00000355693.5 |
COA4
|
cytochrome c oxidase assembly factor 4 homolog |
| chr6_+_167111789 | 0.11 |
ENST00000400926.5
|
CCR6
|
C-C motif chemokine receptor 6 |
| chr11_-_85665077 | 0.11 |
ENST00000527447.2
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr1_+_207496229 | 0.10 |
ENST00000367051.6
ENST00000367053.6 ENST00000367052.6 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr8_-_16186270 | 0.10 |
ENST00000445506.6
|
MSR1
|
macrophage scavenger receptor 1 |
| chr12_-_75209701 | 0.10 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr1_-_51990679 | 0.09 |
ENST00000371655.4
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr7_+_149872955 | 0.09 |
ENST00000421974.7
ENST00000456496.7 |
ATP6V0E2
|
ATPase H+ transporting V0 subunit e2 |
| chr12_-_6470643 | 0.09 |
ENST00000535180.5
ENST00000400911.7 |
VAMP1
|
vesicle associated membrane protein 1 |
| chrM_+_4467 | 0.08 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr16_+_32066065 | 0.08 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chrX_+_71283577 | 0.08 |
ENST00000420903.6
ENST00000373856.8 ENST00000678437.1 ENST00000678830.1 ENST00000677879.1 ENST00000373841.5 ENST00000276079.13 ENST00000676797.1 ENST00000678660.1 ENST00000678231.1 ENST00000677612.1 ENST00000413858.5 ENST00000450092.6 |
NONO
|
non-POU domain containing octamer binding |
| chr1_+_209938207 | 0.08 |
ENST00000472886.5
|
SYT14
|
synaptotagmin 14 |
| chr2_+_113437691 | 0.08 |
ENST00000259199.9
ENST00000416503.6 ENST00000433343.6 |
CBWD2
|
COBW domain containing 2 |
| chr2_+_1413456 | 0.08 |
ENST00000539820.5
ENST00000382269.7 ENST00000345913.8 ENST00000329066.9 ENST00000382201.7 |
TPO
|
thyroid peroxidase |
| chr9_-_4666347 | 0.08 |
ENST00000381890.9
ENST00000682582.1 |
SPATA6L
|
spermatogenesis associated 6 like |
| chrX_-_49080066 | 0.08 |
ENST00000634944.1
ENST00000423215.3 ENST00000465382.6 |
WDR45
|
WD repeat domain 45 |
| chr12_-_11269696 | 0.07 |
ENST00000381842.7
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr11_-_117098415 | 0.07 |
ENST00000445177.6
ENST00000375300.6 ENST00000446921.6 |
SIK3
|
SIK family kinase 3 |
| chr15_+_51751587 | 0.07 |
ENST00000539962.6
ENST00000249700.9 |
TMOD2
|
tropomodulin 2 |
| chr4_-_75724362 | 0.07 |
ENST00000677583.1
|
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr1_-_35031905 | 0.07 |
ENST00000317538.9
ENST00000357182.9 |
ZMYM6
|
zinc finger MYM-type containing 6 |
| chr12_-_62192762 | 0.07 |
ENST00000416284.8
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr6_+_26538338 | 0.07 |
ENST00000377575.3
|
HMGN4
|
high mobility group nucleosomal binding domain 4 |
| chr4_+_70721953 | 0.07 |
ENST00000381006.8
ENST00000226328.8 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr9_+_105662457 | 0.07 |
ENST00000334077.6
|
TAL2
|
TAL bHLH transcription factor 2 |
| chr9_-_110999458 | 0.06 |
ENST00000374430.6
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr16_+_50025217 | 0.06 |
ENST00000427478.7
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr10_-_45535346 | 0.06 |
ENST00000453424.7
ENST00000395769.6 |
MARCHF8
|
membrane associated ring-CH-type finger 8 |
| chr1_+_209938169 | 0.06 |
ENST00000367019.5
ENST00000537238.5 ENST00000637265.1 |
SYT14
|
synaptotagmin 14 |
| chr9_-_101435760 | 0.06 |
ENST00000647789.2
ENST00000616752.1 |
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr1_-_85708382 | 0.05 |
ENST00000370574.4
ENST00000431532.6 |
ZNHIT6
|
zinc finger HIT-type containing 6 |
| chr19_+_16496383 | 0.05 |
ENST00000594035.5
ENST00000221671.8 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr17_+_38297023 | 0.05 |
ENST00000619548.1
ENST00000613675.5 |
MRPL45
|
mitochondrial ribosomal protein L45 |
| chr7_+_20647343 | 0.05 |
ENST00000443026.6
ENST00000406935.5 |
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chr6_+_10694916 | 0.05 |
ENST00000379568.4
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr8_+_103298433 | 0.05 |
ENST00000522566.5
|
FZD6
|
frizzled class receptor 6 |
| chr7_-_105522264 | 0.05 |
ENST00000469408.6
|
PUS7
|
pseudouridine synthase 7 |
| chr3_+_189789672 | 0.05 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr13_-_26221703 | 0.05 |
ENST00000381570.7
ENST00000346166.7 |
RNF6
|
ring finger protein 6 |
| chr11_+_18172837 | 0.05 |
ENST00000314254.3
|
MRGPRX4
|
MAS related GPR family member X4 |
| chr7_-_15561986 | 0.05 |
ENST00000342526.8
|
AGMO
|
alkylglycerol monooxygenase |
| chr13_-_48413105 | 0.04 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr6_-_39322688 | 0.04 |
ENST00000437525.3
|
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr2_+_165294031 | 0.04 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr3_+_141262614 | 0.04 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr4_-_65670339 | 0.04 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5 |
| chr6_-_170553216 | 0.03 |
ENST00000262193.7
|
PSMB1
|
proteasome 20S subunit beta 1 |
| chr1_-_149861210 | 0.03 |
ENST00000579512.2
|
H4C15
|
H4 clustered histone 15 |
| chr8_+_24384275 | 0.03 |
ENST00000256412.8
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr1_-_21783134 | 0.03 |
ENST00000308271.14
|
USP48
|
ubiquitin specific peptidase 48 |
| chr1_-_51878711 | 0.03 |
ENST00000352171.12
|
NRDC
|
nardilysin convertase |
| chr4_-_65670478 | 0.02 |
ENST00000613740.5
ENST00000622150.4 ENST00000511294.1 |
EPHA5
|
EPH receptor A5 |
| chr4_+_15339818 | 0.02 |
ENST00000397700.6
ENST00000295297.4 |
C1QTNF7
|
C1q and TNF related 7 |
| chr17_+_63484840 | 0.02 |
ENST00000290863.10
ENST00000413513.7 |
ACE
|
angiotensin I converting enzyme |
| chrX_-_153926254 | 0.01 |
ENST00000393721.5
ENST00000370028.7 ENST00000350060.10 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr12_-_6470667 | 0.01 |
ENST00000361716.8
ENST00000396308.4 |
VAMP1
|
vesicle associated membrane protein 1 |
| chr3_-_126558926 | 0.01 |
ENST00000318225.3
|
C3orf22
|
chromosome 3 open reading frame 22 |
| chrX_-_140784366 | 0.01 |
ENST00000674533.1
|
CDR1
|
cerebellar degeneration related protein 1 |
| chr1_-_11796536 | 0.01 |
ENST00000641820.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr11_+_65027402 | 0.01 |
ENST00000377244.8
ENST00000534637.5 ENST00000524831.5 |
SNX15
|
sorting nexin 15 |
| chr4_-_47463649 | 0.00 |
ENST00000381571.6
|
COMMD8
|
COMM domain containing 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.5 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.2 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.7 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.6 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.1 | 1.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.5 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.3 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.1 | 0.2 | GO:2000547 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 1.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.0 | 0.3 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.3 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0051586 | peptidyl-cysteine methylation(GO:0018125) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.4 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 3.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.5 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 3.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |