Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for ALX1_ARX
Z-value: 0.46
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
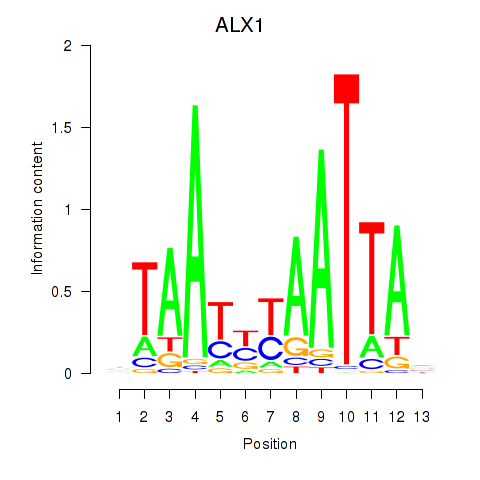
ALX1
|
ENSG00000180318.4 | ALX1 |
|
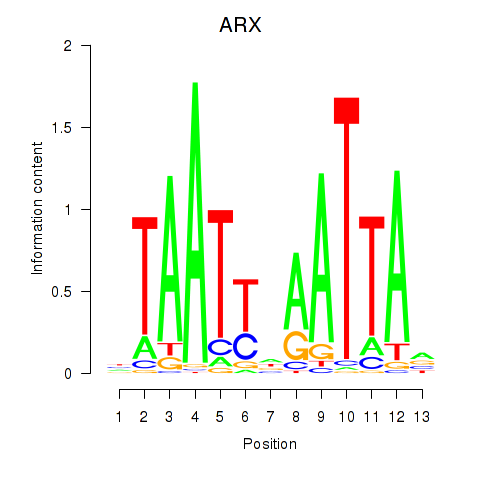
ARX
|
ENSG00000004848.8 | ARX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARX | hg38_v1_chrX_-_25015924_25016002 | -0.42 | 2.1e-02 | Click! |
| ALX1 | hg38_v1_chr12_+_85280199_85280237 | 0.01 | 9.6e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_68447453 | 1.47 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr6_+_34236865 | 1.09 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr6_-_65707214 | 0.94 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chr14_+_23630109 | 0.81 |
ENST00000432832.6
|
DHRS2
|
dehydrogenase/reductase 2 |
| chr6_+_121437378 | 0.78 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr9_+_72577939 | 0.77 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr2_-_31217511 | 0.76 |
ENST00000403897.4
|
CAPN14
|
calpain 14 |
| chr1_+_84164962 | 0.75 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr8_-_90082871 | 0.69 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr3_+_172754457 | 0.69 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr1_+_50103903 | 0.68 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr2_-_55334529 | 0.61 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr5_-_39270623 | 0.55 |
ENST00000512138.1
ENST00000646045.2 |
FYB1
|
FYN binding protein 1 |
| chr3_-_142029108 | 0.55 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr9_+_121567057 | 0.55 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr4_+_168092530 | 0.54 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr1_-_93681829 | 0.51 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr12_-_89352395 | 0.50 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr17_-_62808339 | 0.49 |
ENST00000583600.5
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr17_-_62808299 | 0.48 |
ENST00000311269.10
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr2_+_227813834 | 0.47 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr12_-_89352487 | 0.47 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_84181630 | 0.46 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr12_-_118359639 | 0.45 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr12_+_59689337 | 0.44 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr9_+_72577369 | 0.43 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr7_-_111392915 | 0.43 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr1_+_154429315 | 0.42 |
ENST00000476006.5
|
IL6R
|
interleukin 6 receptor |
| chrX_+_108045050 | 0.39 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_108044967 | 0.39 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr9_+_72577788 | 0.37 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chrX_+_11293411 | 0.36 |
ENST00000348912.4
ENST00000380714.7 ENST00000380712.7 |
AMELX
|
amelogenin X-linked |
| chr10_-_29634964 | 0.36 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr6_-_132659178 | 0.36 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr19_+_7030578 | 0.36 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr19_-_7021431 | 0.35 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr19_-_6393205 | 0.35 |
ENST00000595047.5
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr17_+_76467597 | 0.34 |
ENST00000392492.8
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr15_+_41774459 | 0.32 |
ENST00000457542.7
ENST00000456763.6 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr11_+_66011994 | 0.32 |
ENST00000312134.3
|
CST6
|
cystatin E/M |
| chr19_-_6393131 | 0.32 |
ENST00000394456.10
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr17_-_41118369 | 0.31 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr18_+_616672 | 0.30 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr3_-_125055987 | 0.30 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr18_+_616711 | 0.30 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr6_+_26199509 | 0.29 |
ENST00000356530.5
|
H2BC7
|
H2B clustered histone 7 |
| chr18_-_63644250 | 0.29 |
ENST00000341074.10
ENST00000436264.1 |
SERPINB4
|
serpin family B member 4 |
| chr6_+_63521738 | 0.26 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr4_-_103019634 | 0.26 |
ENST00000510559.1
ENST00000296422.12 ENST00000394789.7 |
SLC9B1
|
solute carrier family 9 member B1 |
| chr2_+_233691607 | 0.26 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr5_+_148312416 | 0.25 |
ENST00000274565.5
|
SPINK7
|
serine peptidase inhibitor Kazal type 7 |
| chr14_-_94770102 | 0.25 |
ENST00000238558.5
|
GSC
|
goosecoid homeobox |
| chr19_-_43465596 | 0.25 |
ENST00000244333.4
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr4_+_118034480 | 0.24 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr18_+_10471829 | 0.24 |
ENST00000584596.2
|
APCDD1
|
APC down-regulated 1 |
| chr3_-_98523013 | 0.23 |
ENST00000394181.6
ENST00000508902.5 ENST00000394180.6 |
CLDND1
|
claudin domain containing 1 |
| chrY_-_6874027 | 0.23 |
ENST00000215479.10
|
AMELY
|
amelogenin Y-linked |
| chr4_-_89029881 | 0.23 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A |
| chr11_+_55811367 | 0.23 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr14_-_80959484 | 0.22 |
ENST00000555529.5
ENST00000556042.5 ENST00000556981.5 |
CEP128
|
centrosomal protein 128 |
| chr9_-_27005659 | 0.21 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr3_-_33645433 | 0.21 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_+_122055355 | 0.21 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chrX_-_11265975 | 0.21 |
ENST00000303025.10
ENST00000657361.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr18_+_63775369 | 0.21 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr4_-_142305935 | 0.21 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr8_-_61689768 | 0.19 |
ENST00000517847.6
ENST00000389204.8 ENST00000517661.5 ENST00000517903.5 ENST00000522603.5 ENST00000541428.5 ENST00000522349.5 ENST00000522835.5 ENST00000518306.5 |
ASPH
|
aspartate beta-hydroxylase |
| chr8_-_10655137 | 0.19 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chr12_-_91153149 | 0.19 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_+_130850585 | 0.19 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr1_-_110391041 | 0.18 |
ENST00000369781.8
ENST00000437429.6 ENST00000541986.5 |
SLC16A4
|
solute carrier family 16 member 4 |
| chr1_-_21050952 | 0.18 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr12_-_11134644 | 0.18 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr1_+_15341744 | 0.18 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr5_+_36151989 | 0.17 |
ENST00000274254.9
|
SKP2
|
S-phase kinase associated protein 2 |
| chr22_-_28306645 | 0.17 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr7_+_107583919 | 0.17 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr3_-_142028597 | 0.17 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chrX_+_120604199 | 0.17 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr2_+_54456311 | 0.17 |
ENST00000615901.4
ENST00000356805.9 |
SPTBN1
|
spectrin beta, non-erythrocytic 1 |
| chr9_+_74615582 | 0.16 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr11_+_89924064 | 0.16 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr12_-_68225806 | 0.16 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr21_+_46635595 | 0.16 |
ENST00000451211.6
ENST00000458387.6 ENST00000397638.6 ENST00000291705.11 ENST00000397637.5 ENST00000334494.8 ENST00000397628.5 ENST00000355680.8 ENST00000440086.5 |
PRMT2
|
protein arginine methyltransferase 2 |
| chr6_+_26087417 | 0.15 |
ENST00000357618.10
ENST00000309234.10 |
HFE
|
homeostatic iron regulator |
| chr12_-_66803980 | 0.15 |
ENST00000539540.5
ENST00000540433.5 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr14_-_81427390 | 0.15 |
ENST00000555447.5
|
STON2
|
stonin 2 |
| chr11_+_121102666 | 0.15 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr1_-_178871060 | 0.15 |
ENST00000234816.7
|
ANGPTL1
|
angiopoietin like 1 |
| chrX_+_120604084 | 0.14 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr12_+_18261511 | 0.14 |
ENST00000538779.6
ENST00000433979.6 |
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr7_-_14841267 | 0.14 |
ENST00000406247.7
ENST00000399322.7 |
DGKB
|
diacylglycerol kinase beta |
| chr5_+_36152077 | 0.14 |
ENST00000546211.6
ENST00000620197.5 ENST00000678270.1 ENST00000679015.1 ENST00000678580.1 ENST00000274255.11 ENST00000508514.5 |
SKP2
|
S-phase kinase associated protein 2 |
| chr11_-_4288083 | 0.14 |
ENST00000638166.1
|
SSU72P4
|
SSU72 pseudogene 4 |
| chr3_-_47892743 | 0.14 |
ENST00000420772.6
|
MAP4
|
microtubule associated protein 4 |
| chr1_+_192158448 | 0.14 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr3_+_12287899 | 0.13 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr5_+_146447304 | 0.13 |
ENST00000296702.9
ENST00000394421.7 ENST00000679501.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr12_-_10172117 | 0.13 |
ENST00000545927.5
ENST00000309539.8 ENST00000432556.6 ENST00000544577.5 |
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr6_-_27867581 | 0.13 |
ENST00000331442.5
|
H1-5
|
H1.5 linker histone, cluster member |
| chr1_+_186828941 | 0.13 |
ENST00000367466.4
|
PLA2G4A
|
phospholipase A2 group IVA |
| chr17_-_41168219 | 0.13 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr12_-_110499546 | 0.13 |
ENST00000552130.6
|
VPS29
|
VPS29 retromer complex component |
| chr10_-_110304894 | 0.13 |
ENST00000369603.10
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr17_-_40867200 | 0.13 |
ENST00000647902.1
ENST00000251643.5 |
KRT12
|
keratin 12 |
| chr13_-_101416441 | 0.12 |
ENST00000675332.1
ENST00000676315.1 ENST00000251127.11 |
NALCN
|
sodium leak channel, non-selective |
| chrX_-_41665766 | 0.12 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr4_+_87832917 | 0.12 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr7_+_136868622 | 0.12 |
ENST00000680005.1
ENST00000445907.6 |
CHRM2
|
cholinergic receptor muscarinic 2 |
| chr11_-_72244166 | 0.12 |
ENST00000298231.5
|
PHOX2A
|
paired like homeobox 2A |
| chr9_-_92536031 | 0.12 |
ENST00000344604.9
ENST00000375540.5 |
ECM2
|
extracellular matrix protein 2 |
| chr4_+_108650644 | 0.12 |
ENST00000512478.2
|
OSTC
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr12_-_6635938 | 0.11 |
ENST00000329858.9
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr6_-_99349647 | 0.11 |
ENST00000389677.6
|
FAXC
|
failed axon connections homolog, metaxin like GST domain containing |
| chr3_+_141262614 | 0.11 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr4_+_108650585 | 0.11 |
ENST00000613215.4
ENST00000361564.9 |
OSTC
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr3_+_12287859 | 0.11 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr3_-_196515315 | 0.11 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chrX_+_38352573 | 0.11 |
ENST00000039007.5
|
OTC
|
ornithine transcarbamylase |
| chr1_+_248095184 | 0.11 |
ENST00000358120.3
ENST00000641893.1 |
OR2L13
|
olfactory receptor family 2 subfamily L member 13 |
| chr3_-_165078480 | 0.11 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr2_+_184598520 | 0.11 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chr3_-_33718207 | 0.11 |
ENST00000359576.9
ENST00000682230.1 ENST00000399362.8 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr20_+_31547367 | 0.11 |
ENST00000394552.3
|
MCTS2P
|
MCTS family member 2, pseudogene |
| chr2_+_118942188 | 0.11 |
ENST00000327097.5
|
MARCO
|
macrophage receptor with collagenous structure |
| chr5_+_141338753 | 0.11 |
ENST00000528330.2
ENST00000394576.3 |
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr11_-_4339244 | 0.10 |
ENST00000524542.2
|
SSU72P7
|
SSU72 pseudogene 7 |
| chr3_+_69936629 | 0.10 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr3_-_87276462 | 0.10 |
ENST00000561167.5
ENST00000560656.1 |
POU1F1
|
POU class 1 homeobox 1 |
| chr3_-_165196689 | 0.10 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr11_-_11353241 | 0.10 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr1_-_109613070 | 0.10 |
ENST00000351050.8
|
GNAT2
|
G protein subunit alpha transducin 2 |
| chr9_-_28670285 | 0.10 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr8_+_38820332 | 0.10 |
ENST00000518809.5
ENST00000520611.1 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr8_-_85341659 | 0.09 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr5_-_9630351 | 0.09 |
ENST00000382492.4
|
TAS2R1
|
taste 2 receptor member 1 |
| chr18_-_5396265 | 0.09 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr2_+_161136901 | 0.09 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr1_+_12857086 | 0.09 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr4_+_155903688 | 0.09 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr12_-_111685720 | 0.08 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr1_-_178871022 | 0.08 |
ENST00000367629.1
|
ANGPTL1
|
angiopoietin like 1 |
| chr12_-_118190510 | 0.08 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr17_-_73232218 | 0.08 |
ENST00000583024.1
ENST00000403627.7 ENST00000405159.7 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104 member A |
| chr6_+_27824084 | 0.08 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr3_-_130746760 | 0.08 |
ENST00000356763.8
|
PIK3R4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr4_-_142305826 | 0.08 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr11_-_13496018 | 0.08 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr11_+_18412292 | 0.08 |
ENST00000541669.6
ENST00000280704.8 |
LDHC
|
lactate dehydrogenase C |
| chr3_-_165196369 | 0.08 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr10_+_68106109 | 0.07 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr2_-_55269038 | 0.07 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr1_+_186296267 | 0.07 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr3_-_39280021 | 0.07 |
ENST00000399220.3
|
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chr3_-_87276577 | 0.07 |
ENST00000344265.8
ENST00000350375.7 |
POU1F1
|
POU class 1 homeobox 1 |
| chr5_+_141364153 | 0.07 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr12_+_12725897 | 0.07 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr5_+_40909490 | 0.07 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr5_+_141364231 | 0.07 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr3_-_9249623 | 0.07 |
ENST00000383836.8
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr14_+_34993240 | 0.06 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr18_-_13915531 | 0.06 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor |
| chr2_+_75646775 | 0.06 |
ENST00000393909.7
ENST00000358788.10 ENST00000409374.5 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr4_+_70195719 | 0.06 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated |
| chr3_+_160225409 | 0.06 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr10_+_27532521 | 0.06 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chr4_-_75724386 | 0.06 |
ENST00000677606.1
ENST00000678798.1 ENST00000677970.1 ENST00000677620.1 ENST00000679281.1 ENST00000677333.1 ENST00000676470.1 ENST00000499709.3 ENST00000511868.6 ENST00000678971.1 ENST00000677265.1 ENST00000677952.1 ENST00000678122.1 ENST00000678100.1 ENST00000678062.1 ENST00000676666.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr3_-_71360753 | 0.06 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1 |
| chr17_-_41098084 | 0.06 |
ENST00000318329.6
ENST00000333822.5 |
KRTAP4-8
|
keratin associated protein 4-8 |
| chr1_+_177170916 | 0.06 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr5_+_173918216 | 0.05 |
ENST00000519467.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr3_+_111674654 | 0.05 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr6_+_132552693 | 0.05 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chr12_-_57479552 | 0.05 |
ENST00000424809.6
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr18_+_68798065 | 0.05 |
ENST00000360242.9
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr8_-_85341705 | 0.05 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr1_+_159008978 | 0.05 |
ENST00000447473.6
|
IFI16
|
interferon gamma inducible protein 16 |
| chr1_+_247549002 | 0.05 |
ENST00000366488.5
|
GCSAML
|
germinal center associated signaling and motility like |
| chr2_+_36696790 | 0.05 |
ENST00000497382.5
ENST00000404084.5 ENST00000379241.7 ENST00000401530.5 |
VIT
|
vitrin |
| chr8_-_121641424 | 0.05 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr10_-_49188380 | 0.05 |
ENST00000374153.7
ENST00000374148.1 ENST00000374151.7 |
TMEM273
|
transmembrane protein 273 |
| chr4_+_154563003 | 0.05 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr12_+_64497968 | 0.04 |
ENST00000676593.1
ENST00000677093.1 |
TBK1
ENSG00000288665.1
|
TANK binding kinase 1 novel transcript |
| chr6_+_30914329 | 0.04 |
ENST00000541562.6
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr6_+_30914205 | 0.04 |
ENST00000672801.1
ENST00000321897.9 ENST00000625423.2 ENST00000676266.1 ENST00000428017.5 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr4_+_94455245 | 0.04 |
ENST00000508216.5
ENST00000514743.5 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr3_-_142028617 | 0.04 |
ENST00000477292.5
ENST00000478006.5 ENST00000495310.5 ENST00000486111.5 |
TFDP2
|
transcription factor Dp-2 |
| chr4_+_75724569 | 0.04 |
ENST00000514213.7
ENST00000264904.8 |
USO1
|
USO1 vesicle transport factor |
| chr12_-_7503744 | 0.04 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr5_+_137867868 | 0.04 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr4_+_87650277 | 0.04 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr2_-_182427014 | 0.04 |
ENST00000409365.5
ENST00000351439.9 |
PDE1A
|
phosphodiesterase 1A |
| chr11_+_85647958 | 0.04 |
ENST00000304511.7
ENST00000528105.5 |
TMEM126A
|
transmembrane protein 126A |
| chr12_-_57479848 | 0.04 |
ENST00000393791.8
ENST00000552249.1 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr11_+_95790459 | 0.03 |
ENST00000325486.9
ENST00000325542.10 ENST00000544522.5 ENST00000541365.5 |
CEP57
|
centrosomal protein 57 |
| chr12_+_71686473 | 0.03 |
ENST00000549735.5
|
TMEM19
|
transmembrane protein 19 |
| chr13_+_53028806 | 0.03 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr4_-_82844418 | 0.03 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr1_-_151175966 | 0.03 |
ENST00000441701.1
ENST00000295314.9 |
TMOD4
|
tropomodulin 4 |
| chr5_+_141343818 | 0.03 |
ENST00000619750.1
ENST00000253812.8 |
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.2 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.8 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 1.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 1.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.5 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.7 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.2 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:0043017 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 0.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.1 | GO:0021623 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.2 | GO:1900390 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 1.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.3 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 1.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.7 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 0.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.7 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |