Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for ARID5A
Z-value: 0.77
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.17 | ARID5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg38_v1_chr2_+_96537254_96537345 | 0.44 | 1.6e-02 | Click! |
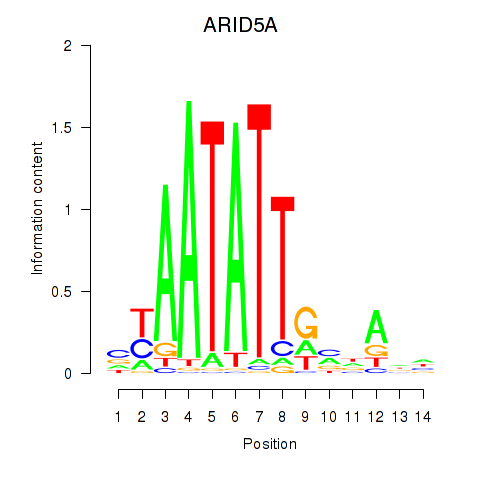
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33283205 | 4.28 |
ENST00000253354.2
|
BPIFB1
|
BPI fold containing family B member 1 |
| chr9_-_34381531 | 3.90 |
ENST00000379124.5
ENST00000379126.7 ENST00000379127.1 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr11_+_62208665 | 3.90 |
ENST00000244930.6
|
SCGB2A1
|
secretoglobin family 2A member 1 |
| chr9_-_34381578 | 3.88 |
ENST00000379133.7
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr8_-_109680812 | 3.35 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr7_-_16881967 | 3.32 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr4_-_99352730 | 3.18 |
ENST00000510055.5
ENST00000515683.6 ENST00000511397.3 |
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide |
| chr13_+_42781578 | 2.90 |
ENST00000313851.3
|
FAM216B
|
family with sequence similarity 216 member B |
| chr13_+_42781547 | 2.89 |
ENST00000537894.5
|
FAM216B
|
family with sequence similarity 216 member B |
| chr5_+_141223332 | 2.64 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chr4_-_176195563 | 2.62 |
ENST00000280191.7
|
SPATA4
|
spermatogenesis associated 4 |
| chr1_+_103617427 | 2.47 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr5_+_140841183 | 2.25 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr10_-_73358709 | 2.24 |
ENST00000340329.7
|
CFAP70
|
cilia and flagella associated protein 70 |
| chr6_+_33080445 | 2.01 |
ENST00000428835.5
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr15_-_52295792 | 2.00 |
ENST00000261839.12
|
MYO5C
|
myosin VC |
| chr16_-_75556214 | 1.97 |
ENST00000568377.5
ENST00000565067.5 ENST00000258173.11 |
TMEM231
|
transmembrane protein 231 |
| chr1_+_78649818 | 1.89 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr6_+_32637419 | 1.88 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr11_-_114595750 | 1.82 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr5_+_140786136 | 1.80 |
ENST00000378133.4
ENST00000504120.4 |
PCDHA1
|
protocadherin alpha 1 |
| chr15_+_70936487 | 1.79 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr11_-_114595777 | 1.73 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr1_+_103750406 | 1.70 |
ENST00000370079.3
|
AMY1C
|
amylase alpha 1C |
| chr3_+_113897470 | 1.60 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr10_+_125973373 | 1.53 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr13_-_51974775 | 1.50 |
ENST00000674147.1
|
ATP7B
|
ATPase copper transporting beta |
| chr19_-_8981342 | 1.46 |
ENST00000397910.8
|
MUC16
|
mucin 16, cell surface associated |
| chr5_+_140882116 | 1.42 |
ENST00000289272.3
ENST00000409494.5 ENST00000617769.1 |
PCDHA13
|
protocadherin alpha 13 |
| chr6_+_33075952 | 1.37 |
ENST00000418931.7
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr1_-_89270751 | 1.30 |
ENST00000370459.7
|
GBP5
|
guanylate binding protein 5 |
| chr6_+_32637396 | 1.26 |
ENST00000395363.5
ENST00000496318.5 ENST00000343139.11 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr11_-_26567087 | 1.23 |
ENST00000436318.6
ENST00000281268.12 |
MUC15
|
mucin 15, cell surface associated |
| chr7_-_120858066 | 1.21 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr15_-_89815332 | 1.18 |
ENST00000559874.2
|
ANPEP
|
alanyl aminopeptidase, membrane |
| chr11_+_17295322 | 1.16 |
ENST00000458064.6
ENST00000622082.4 |
NUCB2
|
nucleobindin 2 |
| chr1_+_170935526 | 1.14 |
ENST00000367758.7
ENST00000367759.9 |
MROH9
|
maestro heat like repeat family member 9 |
| chr5_+_140786291 | 1.14 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1 |
| chr6_+_46793379 | 1.14 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr2_-_32264853 | 1.12 |
ENST00000402280.6
|
NLRC4
|
NLR family CARD domain containing 4 |
| chr3_+_181711915 | 1.10 |
ENST00000325404.3
|
SOX2
|
SRY-box transcription factor 2 |
| chr1_+_47023659 | 1.09 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1 |
| chr4_-_69653223 | 1.04 |
ENST00000286604.8
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus |
| chr12_-_49843092 | 1.02 |
ENST00000333924.6
|
BCDIN3D
|
BCDIN3 domain containing RNA methyltransferase |
| chr1_-_89126066 | 1.00 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr8_-_132625378 | 0.99 |
ENST00000522789.5
|
LRRC6
|
leucine rich repeat containing 6 |
| chr7_-_120858303 | 0.99 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr10_+_94683722 | 0.97 |
ENST00000285979.11
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr10_-_60572599 | 0.96 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr4_+_69096494 | 0.96 |
ENST00000508661.5
ENST00000622664.1 |
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr4_+_69096467 | 0.93 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr12_+_20810698 | 0.92 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr6_-_32668368 | 0.92 |
ENST00000399084.5
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr20_-_18467023 | 0.90 |
ENST00000262547.9
|
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr17_+_70075317 | 0.89 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr5_+_69565122 | 0.89 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr18_+_63907948 | 0.86 |
ENST00000238508.8
|
SERPINB10
|
serpin family B member 10 |
| chr7_+_116953514 | 0.85 |
ENST00000446490.5
|
ST7
|
suppression of tumorigenicity 7 |
| chr6_-_49744434 | 0.85 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr18_-_55351977 | 0.84 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr6_-_49744378 | 0.82 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr11_-_62709493 | 0.82 |
ENST00000405837.5
ENST00000531524.5 ENST00000524862.6 ENST00000679883.1 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr7_+_116953482 | 0.78 |
ENST00000323984.8
ENST00000417919.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_216805367 | 0.78 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr7_+_116953379 | 0.77 |
ENST00000393449.5
|
ST7
|
suppression of tumorigenicity 7 |
| chr7_+_116953238 | 0.75 |
ENST00000393446.6
|
ST7
|
suppression of tumorigenicity 7 |
| chr18_-_55585773 | 0.74 |
ENST00000563824.5
ENST00000626425.2 ENST00000566514.5 ENST00000568673.5 ENST00000562847.5 ENST00000568147.5 |
TCF4
|
transcription factor 4 |
| chr1_+_225825346 | 0.73 |
ENST00000366837.5
|
EPHX1
|
epoxide hydrolase 1 |
| chr16_+_30748241 | 0.72 |
ENST00000565924.5
ENST00000424889.7 |
PHKG2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr5_+_62578810 | 0.71 |
ENST00000334994.6
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr1_+_241532121 | 0.70 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chr18_-_55422306 | 0.70 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr1_-_197067234 | 0.69 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr12_-_14961610 | 0.67 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr12_+_9827517 | 0.67 |
ENST00000537723.5
|
KLRF1
|
killer cell lectin like receptor F1 |
| chr11_-_6030758 | 0.67 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr7_+_117014881 | 0.66 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr8_-_28490220 | 0.65 |
ENST00000517673.5
ENST00000380254.7 ENST00000518734.5 ENST00000346498.6 |
FBXO16
|
F-box protein 16 |
| chr15_-_55365231 | 0.64 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr2_-_40512361 | 0.64 |
ENST00000403092.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr5_-_19988179 | 0.63 |
ENST00000502796.5
ENST00000382275.6 ENST00000511273.1 |
CDH18
|
cadherin 18 |
| chrX_+_56563569 | 0.63 |
ENST00000338222.7
|
UBQLN2
|
ubiquilin 2 |
| chr20_+_15196834 | 0.63 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr12_-_14929116 | 0.62 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr2_-_212124901 | 0.61 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr1_+_40247926 | 0.60 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr3_+_189789672 | 0.60 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr13_-_61415508 | 0.59 |
ENST00000409204.4
|
PCDH20
|
protocadherin 20 |
| chr11_+_60429567 | 0.57 |
ENST00000300190.7
|
MS4A5
|
membrane spanning 4-domains A5 |
| chr2_-_174846405 | 0.57 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr13_-_41194485 | 0.56 |
ENST00000379483.4
|
KBTBD7
|
kelch repeat and BTB domain containing 7 |
| chr8_+_104339796 | 0.56 |
ENST00000622554.1
ENST00000297581.2 |
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr4_+_119135825 | 0.56 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr18_-_55422492 | 0.55 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr3_+_171843337 | 0.53 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr2_-_187448244 | 0.53 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr4_+_55346213 | 0.52 |
ENST00000679836.1
ENST00000264228.9 ENST00000679707.1 |
SRD5A3
ENSG00000288695.1
|
steroid 5 alpha-reductase 3 novel protein, SRD5A3-RP11-177J6.1 readthrough |
| chrX_+_37990773 | 0.51 |
ENST00000341016.5
|
H2AP
|
H2A.P histone |
| chr10_+_68109433 | 0.51 |
ENST00000613327.4
ENST00000358913.10 ENST00000373675.3 |
MYPN
|
myopalladin |
| chr1_-_89065200 | 0.51 |
ENST00000370473.5
|
GBP1
|
guanylate binding protein 1 |
| chr3_+_142623386 | 0.50 |
ENST00000337777.7
ENST00000497199.5 |
PLS1
|
plastin 1 |
| chr11_+_65711991 | 0.50 |
ENST00000377046.7
ENST00000352980.8 |
KAT5
|
lysine acetyltransferase 5 |
| chr2_-_40512423 | 0.50 |
ENST00000402441.5
ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 member A1 |
| chr2_-_174847015 | 0.50 |
ENST00000650938.1
|
CHN1
|
chimerin 1 |
| chr12_-_91179355 | 0.50 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr1_-_160031946 | 0.48 |
ENST00000368090.5
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis class M |
| chr6_-_15548360 | 0.48 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr17_-_75779758 | 0.48 |
ENST00000592643.5
ENST00000591890.5 ENST00000587171.1 ENST00000254810.8 ENST00000589599.5 |
H3-3B
|
H3.3 histone B |
| chr16_+_10386049 | 0.48 |
ENST00000562527.5
ENST00000396559.5 ENST00000396560.6 ENST00000562102.5 ENST00000543967.5 ENST00000569939.5 ENST00000569900.5 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr8_-_93017656 | 0.47 |
ENST00000520686.5
|
TRIQK
|
triple QxxK/R motif containing |
| chr5_+_161685748 | 0.46 |
ENST00000523217.5
|
GABRA6
|
gamma-aminobutyric acid type A receptor subunit alpha6 |
| chr12_+_10307950 | 0.46 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr15_+_65045377 | 0.46 |
ENST00000334287.3
|
SLC51B
|
solute carrier family 51 subunit beta |
| chr1_-_158331522 | 0.45 |
ENST00000368168.4
|
CD1B
|
CD1b molecule |
| chr4_+_183905266 | 0.45 |
ENST00000308497.9
|
STOX2
|
storkhead box 2 |
| chr10_-_125816596 | 0.45 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr1_+_21977014 | 0.45 |
ENST00000337107.11
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr17_+_7252502 | 0.45 |
ENST00000570322.5
ENST00000576496.5 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr5_+_161685708 | 0.44 |
ENST00000274545.10
|
GABRA6
|
gamma-aminobutyric acid type A receptor subunit alpha6 |
| chr18_-_55402187 | 0.44 |
ENST00000630268.2
ENST00000570177.6 |
TCF4
|
transcription factor 4 |
| chrX_+_55717796 | 0.44 |
ENST00000262850.7
|
RRAGB
|
Ras related GTP binding B |
| chr10_+_133394094 | 0.44 |
ENST00000477902.6
|
MTG1
|
mitochondrial ribosome associated GTPase 1 |
| chr1_-_154627945 | 0.43 |
ENST00000681683.1
ENST00000368471.8 ENST00000649042.1 ENST00000680270.1 ENST00000649022.2 ENST00000681056.1 ENST00000649724.1 |
ADAR
|
adenosine deaminase RNA specific |
| chr15_-_55408245 | 0.42 |
ENST00000563171.5
ENST00000425574.7 ENST00000442196.8 ENST00000564092.1 |
CCPG1
|
cell cycle progression 1 |
| chr8_+_74350394 | 0.42 |
ENST00000675928.1
ENST00000676443.1 ENST00000676112.1 ENST00000674710.1 ENST00000434412.3 ENST00000675165.1 ENST00000674973.1 ENST00000220822.12 ENST00000675999.1 ENST00000523640.2 ENST00000524195.2 ENST00000675463.1 ENST00000676207.1 ENST00000674865.1 ENST00000676415.1 |
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr1_-_46176482 | 0.42 |
ENST00000540385.2
ENST00000506599.2 |
P3R3URF-PIK3R3
P3R3URF
|
P3R3URF-PIK3R3 readthrough PIK3R3 upstream reading frame |
| chr6_+_156776020 | 0.41 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B |
| chr7_-_6059139 | 0.41 |
ENST00000446699.1
ENST00000199389.11 |
EIF2AK1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr20_+_6006039 | 0.41 |
ENST00000452938.5
ENST00000378863.9 |
CRLS1
|
cardiolipin synthase 1 |
| chr5_+_126423363 | 0.41 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chr4_+_6269831 | 0.40 |
ENST00000503569.5
ENST00000673991.1 ENST00000682275.1 ENST00000226760.5 |
WFS1
|
wolframin ER transmembrane glycoprotein |
| chr6_-_26271815 | 0.40 |
ENST00000614378.1
|
H3C8
|
H3 clustered histone 8 |
| chr2_+_10368764 | 0.40 |
ENST00000620771.4
|
HPCAL1
|
hippocalcin like 1 |
| chr16_+_53099100 | 0.39 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_-_30992665 | 0.39 |
ENST00000406071.6
|
DCDC1
|
doublecortin domain containing 1 |
| chr10_+_110005804 | 0.39 |
ENST00000360162.7
|
ADD3
|
adducin 3 |
| chr2_-_68952880 | 0.39 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr6_+_83067655 | 0.38 |
ENST00000237163.9
ENST00000349129.7 |
DOP1A
|
DOP1 leucine zipper like protein A |
| chr6_-_25874212 | 0.38 |
ENST00000361703.10
ENST00000397060.8 |
SLC17A3
|
solute carrier family 17 member 3 |
| chrX_+_77899440 | 0.37 |
ENST00000373335.4
ENST00000475465.1 ENST00000650309.2 ENST00000647835.1 |
COX7B
|
cytochrome c oxidase subunit 7B |
| chr12_+_130953898 | 0.37 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr12_+_11171166 | 0.37 |
ENST00000622602.2
|
SMIM10L1
|
small integral membrane protein 10 like 1 |
| chrX_+_55717733 | 0.37 |
ENST00000414239.5
ENST00000374941.9 |
RRAGB
|
Ras related GTP binding B |
| chr2_-_10447771 | 0.37 |
ENST00000405333.5
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr8_+_85177225 | 0.36 |
ENST00000418930.6
|
E2F5
|
E2F transcription factor 5 |
| chr16_+_67570741 | 0.35 |
ENST00000644753.1
ENST00000642819.1 ENST00000645306.1 |
CTCF
|
CCCTC-binding factor |
| chr4_+_6269869 | 0.35 |
ENST00000506362.2
|
WFS1
|
wolframin ER transmembrane glycoprotein |
| chr11_+_112175526 | 0.35 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr6_+_28267107 | 0.35 |
ENST00000621053.1
ENST00000617168.4 ENST00000421553.7 ENST00000611552.2 ENST00000623276.3 |
ENSG00000276302.1
ZSCAN26
|
novel protein zinc finger and SCAN domain containing 26 |
| chr6_+_28267355 | 0.35 |
ENST00000614088.1
ENST00000619937.4 |
ZSCAN26
|
zinc finger and SCAN domain containing 26 |
| chr1_-_169734064 | 0.34 |
ENST00000333360.12
|
SELE
|
selectin E |
| chr12_-_47705990 | 0.34 |
ENST00000432584.7
ENST00000005386.8 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr18_-_63661884 | 0.33 |
ENST00000332821.8
ENST00000283752.10 |
SERPINB3
|
serpin family B member 3 |
| chr2_-_190250503 | 0.33 |
ENST00000409820.2
ENST00000410045.5 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr7_-_32071397 | 0.33 |
ENST00000396184.7
ENST00000396189.2 ENST00000321453.12 |
PDE1C
|
phosphodiesterase 1C |
| chr21_+_44600597 | 0.33 |
ENST00000609664.2
|
KRTAP10-7
|
keratin associated protein 10-7 |
| chr7_-_64982021 | 0.33 |
ENST00000610793.1
ENST00000620222.4 |
ZNF117
|
zinc finger protein 117 |
| chr6_-_76072654 | 0.32 |
ENST00000369950.8
ENST00000611179.4 ENST00000369963.5 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr17_-_45262084 | 0.32 |
ENST00000331780.5
|
SPATA32
|
spermatogenesis associated 32 |
| chr2_+_79025696 | 0.32 |
ENST00000272324.10
|
REG3G
|
regenerating family member 3 gamma |
| chr15_-_77083925 | 0.32 |
ENST00000558745.5
|
TSPAN3
|
tetraspanin 3 |
| chrX_-_10677720 | 0.32 |
ENST00000453318.6
|
MID1
|
midline 1 |
| chr8_+_24294107 | 0.31 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr12_-_47705971 | 0.31 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr13_-_41132728 | 0.31 |
ENST00000379485.2
|
KBTBD6
|
kelch repeat and BTB domain containing 6 |
| chr11_+_22666604 | 0.31 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr3_+_2892199 | 0.30 |
ENST00000397459.6
|
CNTN4
|
contactin 4 |
| chr7_+_116953306 | 0.30 |
ENST00000265437.9
ENST00000393451.7 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_154627906 | 0.30 |
ENST00000679899.1
|
ADAR
|
adenosine deaminase RNA specific |
| chr2_+_79025678 | 0.30 |
ENST00000393897.6
|
REG3G
|
regenerating family member 3 gamma |
| chr17_-_39688016 | 0.29 |
ENST00000579146.5
ENST00000300658.9 ENST00000378011.8 ENST00000429199.6 |
PGAP3
|
post-GPI attachment to proteins phospholipase 3 |
| chr9_+_113349514 | 0.29 |
ENST00000374183.5
|
BSPRY
|
B-box and SPRY domain containing |
| chr17_+_6651745 | 0.29 |
ENST00000542475.3
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr12_-_8066331 | 0.29 |
ENST00000546241.1
ENST00000307637.5 |
C3AR1
|
complement C3a receptor 1 |
| chr10_+_96000091 | 0.28 |
ENST00000424464.5
ENST00000410012.6 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr4_+_109912877 | 0.28 |
ENST00000265171.10
ENST00000509793.5 ENST00000652245.1 |
EGF
|
epidermal growth factor |
| chr10_-_15860450 | 0.28 |
ENST00000277632.8
|
MINDY3
|
MINDY lysine 48 deubiquitinase 3 |
| chr19_-_57477503 | 0.28 |
ENST00000596831.1
ENST00000601768.1 ENST00000600175.5 ENST00000356584.8 ENST00000425074.3 ENST00000343280.8 ENST00000427512.6 ENST00000615214.3 ENST00000610548.2 |
ENSG00000268163.1
ZNF772
|
novel transcript zinc finger protein 772 |
| chr7_-_16645728 | 0.28 |
ENST00000306999.7
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2 |
| chr17_+_80544817 | 0.27 |
ENST00000306801.8
ENST00000570891.5 |
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr20_-_56525925 | 0.27 |
ENST00000243913.8
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr6_-_26056460 | 0.27 |
ENST00000343677.4
|
H1-2
|
H1.2 linker histone, cluster member |
| chrX_+_83861126 | 0.27 |
ENST00000621735.4
ENST00000329312.5 |
CYLC1
|
cylicin 1 |
| chr14_+_67720842 | 0.26 |
ENST00000267502.3
|
RDH12
|
retinol dehydrogenase 12 |
| chr11_+_99020940 | 0.26 |
ENST00000524871.6
|
CNTN5
|
contactin 5 |
| chr6_+_158536398 | 0.25 |
ENST00000367090.4
|
TMEM181
|
transmembrane protein 181 |
| chr6_-_26199272 | 0.25 |
ENST00000650491.1
ENST00000635200.1 ENST00000341023.2 |
ENSG00000282988.2
H2AC7
|
novel protein H2A clustered histone 7 |
| chr5_+_140855882 | 0.25 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr18_+_58362467 | 0.25 |
ENST00000675101.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr13_+_49110309 | 0.25 |
ENST00000398316.7
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr19_+_54502867 | 0.25 |
ENST00000351841.2
|
LAIR2
|
leukocyte associated immunoglobulin like receptor 2 |
| chr5_+_141121793 | 0.24 |
ENST00000194152.4
|
PCDHB4
|
protocadherin beta 4 |
| chr14_+_39233908 | 0.24 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr3_-_48898813 | 0.24 |
ENST00000319017.5
ENST00000430379.5 |
SLC25A20
|
solute carrier family 25 member 20 |
| chr1_-_165698863 | 0.24 |
ENST00000354775.4
|
ALDH9A1
|
aldehyde dehydrogenase 9 family member A1 |
| chr19_-_42442938 | 0.24 |
ENST00000601181.6
|
CXCL17
|
C-X-C motif chemokine ligand 17 |
| chr19_+_45995445 | 0.23 |
ENST00000536603.5
ENST00000595358.5 ENST00000594672.5 |
CCDC61
|
coiled-coil domain containing 61 |
| chr18_+_6834473 | 0.23 |
ENST00000581099.5
ENST00000419673.6 ENST00000531294.5 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr3_+_101561833 | 0.23 |
ENST00000309922.7
|
TRMT10C
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr10_+_133394119 | 0.23 |
ENST00000317502.11
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome associated GTPase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0060003 | copper ion export(GO:0060003) |
| 0.4 | 2.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 4.3 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 0.7 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 0.2 | 1.0 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.6 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.2 | 3.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 0.6 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 0.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 1.3 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.2 | 1.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 0.6 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 0.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.6 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 0.4 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 2.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.7 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.7 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.1 | 0.5 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 1.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 1.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 1.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.8 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0000964 | mitochondrial RNA 5'-end processing(GO:0000964) |
| 0.1 | 0.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 1.0 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 2.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.4 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 7.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.3 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.2 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.4 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 8.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.5 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:1990418 | response to sodium phosphate(GO:1904383) response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 4.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 3.4 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 0.2 | GO:0071033 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 2.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.4 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:2000836 | androgen secretion(GO:0035935) testosterone secretion(GO:0035936) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.8 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 1.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.4 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.5 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 3.4 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 0.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.3 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 0.3 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.1 | 2.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 1.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 2.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.5 | 1.5 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.4 | 3.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 4.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 3.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.5 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 1.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.4 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.3 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 2.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 3.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 2.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.3 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 2.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 0.1 | 2.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0019534 | efflux transmembrane transporter activity(GO:0015562) toxin transporter activity(GO:0019534) |
| 0.1 | 0.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.5 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 6.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 3.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 2.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |