Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
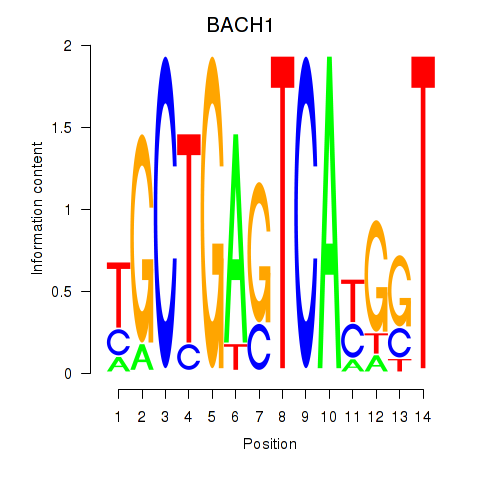
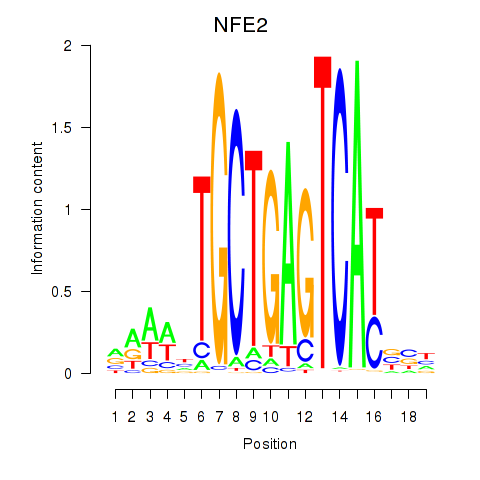
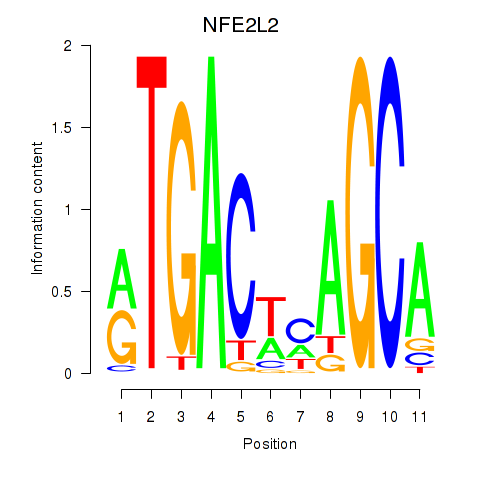
Results for BACH1_NFE2_NFE2L2
Z-value: 0.69
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.16 | BACH1 |
|
NFE2
|
ENSG00000123405.14 | NFE2 |
|
NFE2L2
|
ENSG00000116044.16 | NFE2L2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BACH1 | hg38_v1_chr21_+_29298890_29298932, hg38_v1_chr21_+_29299368_29299441 | -0.23 | 2.1e-01 | Click! |
| NFE2 | hg38_v1_chr12_-_54300974_54301021, hg38_v1_chr12_-_54295748_54295798 | 0.20 | 2.9e-01 | Click! |
| NFE2L2 | hg38_v1_chr2_-_177392673_177392704 | -0.17 | 3.7e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_132066003 | 2.06 |
ENST00000657318.1
ENST00000666210.1 |
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr10_+_132065937 | 2.03 |
ENST00000658847.1
ENST00000666974.1 |
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr12_+_55681647 | 1.93 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B |
| chr12_+_55681711 | 1.89 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr22_+_19479457 | 1.40 |
ENST00000407835.6
ENST00000455750.6 |
CDC45
|
cell division cycle 45 |
| chr1_+_206770764 | 0.80 |
ENST00000656872.2
ENST00000659997.2 |
IL19
|
interleukin 19 |
| chr10_+_133087883 | 0.68 |
ENST00000392607.8
|
ADGRA1
|
adhesion G protein-coupled receptor A1 |
| chr17_+_32444379 | 0.61 |
ENST00000578213.5
ENST00000649012.1 ENST00000457654.6 ENST00000579451.1 ENST00000261712.8 |
PSMD11
|
proteasome 26S subunit, non-ATPase 11 |
| chr8_+_74339566 | 0.60 |
ENST00000675376.1
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr10_-_125816510 | 0.56 |
ENST00000650587.1
|
UROS
|
uroporphyrinogen III synthase |
| chr1_-_235588959 | 0.53 |
ENST00000484517.2
|
GNG4
|
G protein subunit gamma 4 |
| chr14_-_23155302 | 0.52 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr2_+_10123171 | 0.52 |
ENST00000615152.5
|
RRM2
|
ribonucleotide reductase regulatory subunit M2 |
| chr14_-_64942783 | 0.50 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr12_-_70788914 | 0.49 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr10_-_125816596 | 0.49 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr14_-_64942720 | 0.49 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chrX_+_154542194 | 0.47 |
ENST00000618670.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr19_+_46303599 | 0.47 |
ENST00000300862.7
|
HIF3A
|
hypoxia inducible factor 3 subunit alpha |
| chr19_+_55836532 | 0.46 |
ENST00000301295.11
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chr19_+_50203607 | 0.42 |
ENST00000642316.2
ENST00000425460.6 ENST00000440075.6 ENST00000376970.6 ENST00000599920.5 |
MYH14
|
myosin heavy chain 14 |
| chr16_+_89921851 | 0.41 |
ENST00000554444.5
ENST00000556565.5 |
TUBB3
|
tubulin beta 3 class III |
| chr2_-_210315160 | 0.40 |
ENST00000352451.4
|
MYL1
|
myosin light chain 1 |
| chr16_-_69726506 | 0.40 |
ENST00000561500.5
ENST00000320623.10 ENST00000439109.6 ENST00000564043.1 ENST00000379046.6 ENST00000379047.7 |
NQO1
|
NAD(P)H quinone dehydrogenase 1 |
| chr14_+_58244821 | 0.40 |
ENST00000216455.9
ENST00000412908.6 ENST00000557508.5 |
PSMA3
|
proteasome 20S subunit alpha 3 |
| chr18_+_158513 | 0.39 |
ENST00000400266.7
ENST00000580410.5 ENST00000261601.8 ENST00000383589.6 |
USP14
|
ubiquitin specific peptidase 14 |
| chr4_+_4387078 | 0.38 |
ENST00000504171.1
|
NSG1
|
neuronal vesicle trafficking associated 1 |
| chr6_-_83431038 | 0.38 |
ENST00000369705.4
|
ME1
|
malic enzyme 1 |
| chr3_-_170870033 | 0.36 |
ENST00000466674.5
ENST00000295830.13 ENST00000463836.1 |
RPL22L1
|
ribosomal protein L22 like 1 |
| chr1_-_150235972 | 0.34 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr3_-_48016400 | 0.33 |
ENST00000434267.5
ENST00000683076.1 ENST00000633710.1 |
MAP4
|
microtubule associated protein 4 |
| chr4_+_4387039 | 0.32 |
ENST00000621129.4
|
NSG1
|
neuronal vesicle trafficking associated 1 |
| chr22_+_44677077 | 0.32 |
ENST00000403581.5
|
PRR5
|
proline rich 5 |
| chrX_-_73214793 | 0.31 |
ENST00000373517.4
|
NAP1L2
|
nucleosome assembly protein 1 like 2 |
| chr22_+_44677044 | 0.31 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chr5_-_138575359 | 0.31 |
ENST00000297185.9
ENST00000678300.1 ENST00000677425.1 ENST00000677064.1 ENST00000507115.6 |
HSPA9
|
heat shock protein family A (Hsp70) member 9 |
| chr19_-_15934521 | 0.30 |
ENST00000402119.9
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr6_+_86937500 | 0.30 |
ENST00000305344.7
|
HTR1E
|
5-hydroxytryptamine receptor 1E |
| chr22_+_44676808 | 0.29 |
ENST00000624862.3
|
PRR5
|
proline rich 5 |
| chr17_-_75154534 | 0.29 |
ENST00000356033.8
|
JPT1
|
Jupiter microtubule associated homolog 1 |
| chr15_-_82647960 | 0.29 |
ENST00000615198.4
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr16_+_83953232 | 0.29 |
ENST00000565123.5
ENST00000393306.6 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr1_-_93909329 | 0.29 |
ENST00000370238.8
ENST00000615724.1 |
GCLM
|
glutamate-cysteine ligase modifier subunit |
| chr22_+_22697789 | 0.28 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr9_+_72577939 | 0.28 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr14_+_21990357 | 0.28 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr17_-_15566276 | 0.27 |
ENST00000630868.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr2_-_219254558 | 0.27 |
ENST00000392088.6
|
TUBA4A
|
tubulin alpha 4a |
| chr7_-_1160144 | 0.26 |
ENST00000397083.6
ENST00000401903.5 ENST00000316495.8 |
ZFAND2A
|
zinc finger AN1-type containing 2A |
| chr16_-_30113528 | 0.26 |
ENST00000406256.8
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr10_-_50279715 | 0.26 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chr17_-_40703744 | 0.26 |
ENST00000264651.3
|
KRT24
|
keratin 24 |
| chr18_+_31318144 | 0.25 |
ENST00000257192.5
|
DSG1
|
desmoglein 1 |
| chr19_-_15934853 | 0.25 |
ENST00000620614.4
ENST00000248041.12 |
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr11_+_6845683 | 0.25 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr1_-_201023694 | 0.24 |
ENST00000332129.6
ENST00000422435.2 ENST00000461742.7 |
KIF21B
|
kinesin family member 21B |
| chr8_+_86098901 | 0.24 |
ENST00000285393.4
|
ATP6V0D2
|
ATPase H+ transporting V0 subunit d2 |
| chr14_-_106088573 | 0.24 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr22_+_22327298 | 0.24 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr9_-_110256466 | 0.24 |
ENST00000374515.9
ENST00000374517.6 |
TXN
|
thioredoxin |
| chr22_+_22357739 | 0.24 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr3_+_46163465 | 0.23 |
ENST00000357422.2
|
CCR3
|
C-C motif chemokine receptor 3 |
| chr15_-_82571803 | 0.23 |
ENST00000617462.4
ENST00000618449.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr21_+_32298945 | 0.22 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr10_-_97292625 | 0.21 |
ENST00000466484.1
ENST00000358531.9 ENST00000358308.7 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr18_+_63970029 | 0.21 |
ENST00000353706.6
ENST00000542677.5 ENST00000397985.7 ENST00000636430.1 ENST00000397988.7 ENST00000448851.5 |
SERPINB8
|
serpin family B member 8 |
| chr19_-_58511981 | 0.21 |
ENST00000263093.7
ENST00000601355.1 |
SLC27A5
|
solute carrier family 27 member 5 |
| chr2_+_89851723 | 0.20 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr6_-_105137147 | 0.20 |
ENST00000314641.10
|
BVES
|
blood vessel epicardial substance |
| chr3_+_184319677 | 0.20 |
ENST00000441154.5
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr19_-_49155130 | 0.20 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chr1_+_173868076 | 0.20 |
ENST00000367704.5
|
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr2_-_88992903 | 0.20 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr17_-_41612757 | 0.20 |
ENST00000301653.9
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr3_+_184320283 | 0.20 |
ENST00000428387.5
ENST00000434061.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr14_+_21749163 | 0.20 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr19_-_49155384 | 0.19 |
ENST00000252825.9
|
HRC
|
histidine rich calcium binding protein |
| chr4_-_993376 | 0.19 |
ENST00000398520.6
ENST00000398516.3 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr17_-_4545065 | 0.18 |
ENST00000572759.1
|
MYBBP1A
|
MYB binding protein 1a |
| chr11_+_123590939 | 0.18 |
ENST00000646146.1
|
GRAMD1B
|
GRAM domain containing 1B |
| chr2_-_42764116 | 0.18 |
ENST00000378661.3
|
OXER1
|
oxoeicosanoid receptor 1 |
| chr22_+_22734577 | 0.18 |
ENST00000390310.3
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr7_+_141764097 | 0.18 |
ENST00000247879.2
|
TAS2R3
|
taste 2 receptor member 3 |
| chr2_-_169031317 | 0.18 |
ENST00000650372.1
|
ABCB11
|
ATP binding cassette subfamily B member 11 |
| chr17_-_78925376 | 0.18 |
ENST00000262768.11
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr1_-_162868761 | 0.18 |
ENST00000367910.5
ENST00000367912.6 |
CCDC190
|
coiled-coil domain containing 190 |
| chr15_-_76012390 | 0.18 |
ENST00000394907.8
|
NRG4
|
neuregulin 4 |
| chr17_-_10838075 | 0.18 |
ENST00000580256.3
ENST00000643787.1 |
PIRT
ENSG00000284876.1
|
phosphoinositide interacting regulator of transient receptor potential channels novel transcript |
| chr11_-_123885627 | 0.17 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chr17_-_5234801 | 0.17 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr1_-_225428813 | 0.17 |
ENST00000338179.6
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr17_-_41620801 | 0.17 |
ENST00000648859.1
|
KRT17
|
keratin 17 |
| chr6_-_11807045 | 0.16 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr12_+_75480745 | 0.16 |
ENST00000266659.8
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr14_-_24263162 | 0.16 |
ENST00000206765.11
ENST00000544573.5 |
TGM1
|
transglutaminase 1 |
| chr5_+_179820895 | 0.16 |
ENST00000504627.1
ENST00000389805.9 ENST00000510187.5 |
SQSTM1
|
sequestosome 1 |
| chr4_-_79408198 | 0.16 |
ENST00000358842.5
|
GK2
|
glycerol kinase 2 |
| chr17_-_42181081 | 0.15 |
ENST00000607371.5
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chr12_+_75480800 | 0.15 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr20_-_653189 | 0.15 |
ENST00000381962.4
|
SRXN1
|
sulfiredoxin 1 |
| chr6_+_47698538 | 0.15 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr8_-_30727777 | 0.15 |
ENST00000537535.5
ENST00000541648.5 ENST00000546342.5 ENST00000221130.11 |
GSR
|
glutathione-disulfide reductase |
| chr16_+_57628684 | 0.15 |
ENST00000567397.5
ENST00000568979.5 ENST00000672974.1 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr19_+_18588789 | 0.15 |
ENST00000450195.6
ENST00000358607.11 |
REX1BD
|
required for excision 1-B domain containing |
| chr1_+_84144260 | 0.15 |
ENST00000370685.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr6_+_47698574 | 0.15 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr17_-_42181116 | 0.15 |
ENST00000264661.4
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chr12_-_86838867 | 0.15 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C |
| chr11_+_10456186 | 0.15 |
ENST00000528723.5
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr17_+_4950147 | 0.15 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr9_-_137188540 | 0.15 |
ENST00000323927.3
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr2_-_186849164 | 0.15 |
ENST00000295131.3
|
ZSWIM2
|
zinc finger SWIM-type containing 2 |
| chr22_+_22704265 | 0.14 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr1_+_75786246 | 0.14 |
ENST00000319942.8
|
RABGGTB
|
Rab geranylgeranyltransferase subunit beta |
| chr12_-_121016345 | 0.14 |
ENST00000535367.1
ENST00000538296.5 ENST00000288757.7 ENST00000539736.5 ENST00000537817.5 |
C12orf43
|
chromosome 12 open reading frame 43 |
| chr12_-_14961610 | 0.14 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr1_+_152663378 | 0.14 |
ENST00000368784.2
|
LCE2D
|
late cornified envelope 2D |
| chr9_-_26947222 | 0.14 |
ENST00000520884.5
ENST00000397292.8 |
PLAA
|
phospholipase A2 activating protein |
| chr11_+_119087979 | 0.14 |
ENST00000535253.5
ENST00000392841.1 ENST00000648374.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr2_+_234050732 | 0.14 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2 |
| chr2_+_231056845 | 0.14 |
ENST00000677230.1
ENST00000677259.1 ENST00000677180.1 ENST00000409643.6 ENST00000619128.5 ENST00000678679.1 ENST00000676740.1 ENST00000308696.11 ENST00000440838.5 ENST00000373635.9 |
PSMD1
|
proteasome 26S subunit, non-ATPase 1 |
| chr15_+_65045377 | 0.14 |
ENST00000334287.3
|
SLC51B
|
solute carrier family 51 subunit beta |
| chr6_+_44247866 | 0.13 |
ENST00000371554.2
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr6_+_44247087 | 0.13 |
ENST00000353801.7
ENST00000371646.10 |
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr1_+_160190567 | 0.13 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr3_+_184299198 | 0.13 |
ENST00000417952.5
ENST00000310118.9 |
PSMD2
|
proteasome 26S subunit, non-ATPase 2 |
| chr9_-_35072561 | 0.13 |
ENST00000678650.1
|
VCP
|
valosin containing protein |
| chr16_+_22297375 | 0.13 |
ENST00000615879.4
ENST00000299853.10 ENST00000564209.5 ENST00000565358.5 ENST00000418581.6 ENST00000564883.5 ENST00000359210.8 ENST00000563024.5 |
POLR3E
|
RNA polymerase III subunit E |
| chr1_+_152675295 | 0.13 |
ENST00000368783.1
|
LCE2C
|
late cornified envelope 2C |
| chr2_+_17878637 | 0.12 |
ENST00000304101.9
|
KCNS3
|
potassium voltage-gated channel modifier subfamily S member 3 |
| chr17_+_2594148 | 0.12 |
ENST00000675331.1
|
PAFAH1B1
|
platelet activating factor acetylhydrolase 1b regulatory subunit 1 |
| chr17_+_2593925 | 0.12 |
ENST00000674717.1
ENST00000676353.1 ENST00000675202.1 ENST00000674608.1 ENST00000676098.1 |
PAFAH1B1
|
platelet activating factor acetylhydrolase 1b regulatory subunit 1 |
| chr12_-_14961256 | 0.12 |
ENST00000541380.5
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr7_-_116030750 | 0.12 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr2_-_219308963 | 0.12 |
ENST00000423636.6
ENST00000442029.5 ENST00000412847.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr5_-_150155828 | 0.12 |
ENST00000261799.9
|
PDGFRB
|
platelet derived growth factor receptor beta |
| chr17_-_63827647 | 0.12 |
ENST00000584574.5
ENST00000585145.1 ENST00000427159.7 |
FTSJ3
|
FtsJ RNA 2'-O-methyltransferase 3 |
| chr14_-_106622837 | 0.12 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr8_-_143939543 | 0.12 |
ENST00000345136.8
|
PLEC
|
plectin |
| chr14_-_105987068 | 0.12 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr15_+_41559189 | 0.12 |
ENST00000263798.8
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr16_+_57628507 | 0.12 |
ENST00000456916.5
ENST00000567154.5 ENST00000388813.9 ENST00000562558.6 ENST00000566271.6 ENST00000563374.5 ENST00000673126.2 ENST00000562631.7 ENST00000568234.5 ENST00000565770.5 ENST00000566164.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr16_+_78022538 | 0.11 |
ENST00000651443.1
ENST00000299642.10 |
CLEC3A
|
C-type lectin domain family 3 member A |
| chr2_-_152098810 | 0.11 |
ENST00000636442.1
ENST00000638005.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr5_-_177496845 | 0.11 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr1_-_46132650 | 0.11 |
ENST00000372006.5
ENST00000425892.2 ENST00000420542.5 |
PIK3R3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chrX_-_57680260 | 0.11 |
ENST00000434992.1
|
NLRP2B
|
NLR family pyrin domain containing 2B |
| chr22_+_22395005 | 0.11 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr18_-_57586668 | 0.11 |
ENST00000592699.6
ENST00000382873.8 ENST00000262093.11 ENST00000652755.1 |
FECH
|
ferrochelatase |
| chr17_-_60392333 | 0.11 |
ENST00000590133.5
|
USP32
|
ubiquitin specific peptidase 32 |
| chr18_-_37485747 | 0.11 |
ENST00000589229.5
ENST00000587819.5 |
CELF4
|
CUGBP Elav-like family member 4 |
| chr10_+_69630227 | 0.11 |
ENST00000373279.6
|
FAM241B
|
family with sequence similarity 241 member B |
| chr5_+_161848112 | 0.11 |
ENST00000393943.10
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr19_+_8832398 | 0.11 |
ENST00000595891.1
|
MBD3L1
|
methyl-CpG binding domain protein 3 like 1 |
| chr12_-_52949818 | 0.11 |
ENST00000546897.5
ENST00000552551.5 |
KRT8
|
keratin 8 |
| chr4_-_38856807 | 0.11 |
ENST00000506146.5
ENST00000436693.6 ENST00000508254.5 ENST00000514655.1 |
TLR1
TLR6
|
toll like receptor 1 toll like receptor 6 |
| chr3_-_149576203 | 0.11 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr1_-_161307420 | 0.10 |
ENST00000491222.5
ENST00000672287.2 |
MPZ
|
myelin protein zero |
| chr2_+_171999937 | 0.10 |
ENST00000315796.5
|
METAP1D
|
methionyl aminopeptidase type 1D, mitochondrial |
| chr12_+_75334675 | 0.10 |
ENST00000312442.2
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr11_-_2903490 | 0.10 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chr17_-_58531941 | 0.10 |
ENST00000581607.1
ENST00000317256.10 ENST00000426861.5 ENST00000580809.5 ENST00000577729.5 ENST00000583291.1 |
SEPTIN4
|
septin 4 |
| chrX_-_153944621 | 0.10 |
ENST00000393700.8
|
RENBP
|
renin binding protein |
| chr1_+_43337803 | 0.10 |
ENST00000372470.9
ENST00000413998.7 |
MPL
|
MPL proto-oncogene, thrombopoietin receptor |
| chr22_-_45212431 | 0.10 |
ENST00000496226.1
ENST00000251993.11 |
KIAA0930
|
KIAA0930 |
| chr9_-_127874964 | 0.10 |
ENST00000373156.5
|
AK1
|
adenylate kinase 1 |
| chr12_+_75334655 | 0.10 |
ENST00000378695.9
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr7_+_103347635 | 0.10 |
ENST00000679250.1
ENST00000292644.5 ENST00000425206.6 |
PSMC2
|
proteasome 26S subunit, ATPase 2 |
| chrX_-_20141810 | 0.10 |
ENST00000379593.1
ENST00000379607.10 |
EIF1AX
|
eukaryotic translation initiation factor 1A X-linked |
| chr22_+_22720615 | 0.10 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr14_+_21918161 | 0.10 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr8_-_93741001 | 0.10 |
ENST00000518597.2
ENST00000520560.6 ENST00000399300.7 |
RBM12B
|
RNA binding motif protein 12B |
| chr12_-_55707865 | 0.10 |
ENST00000347027.10
ENST00000257879.11 ENST00000553804.6 |
ITGA7
|
integrin subunit alpha 7 |
| chr8_-_93740975 | 0.10 |
ENST00000517700.6
|
RBM12B
|
RNA binding motif protein 12B |
| chrX_+_150363306 | 0.10 |
ENST00000370401.7
ENST00000432680.7 |
MAMLD1
|
mastermind like domain containing 1 |
| chr5_-_177496802 | 0.10 |
ENST00000506161.5
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr6_-_32407123 | 0.09 |
ENST00000374993.4
ENST00000544175.2 ENST00000454136.7 ENST00000446536.2 |
BTNL2
|
butyrophilin like 2 |
| chr12_+_50400809 | 0.09 |
ENST00000293618.12
ENST00000429001.7 ENST00000398473.7 ENST00000548174.5 ENST00000548697.5 ENST00000548993.5 ENST00000614335.4 ENST00000522085.5 ENST00000615080.4 ENST00000518444.5 ENST00000551886.5 ENST00000523389.5 ENST00000518561.5 ENST00000347328.9 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein 4 |
| chr9_+_35538619 | 0.09 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr12_-_57632668 | 0.09 |
ENST00000552350.5
ENST00000548888.5 |
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyltransferase 1 |
| chrX_+_150363258 | 0.09 |
ENST00000683696.1
|
MAMLD1
|
mastermind like domain containing 1 |
| chr19_-_45639104 | 0.09 |
ENST00000586770.5
ENST00000591721.5 ENST00000245925.8 ENST00000590043.5 ENST00000589876.5 |
EML2
|
EMAP like 2 |
| chr19_-_6670117 | 0.09 |
ENST00000245912.7
|
TNFSF14
|
TNF superfamily member 14 |
| chr7_-_100428657 | 0.09 |
ENST00000360951.8
ENST00000398027.6 ENST00000684423.1 ENST00000472716.1 |
ZCWPW1
|
zinc finger CW-type and PWWP domain containing 1 |
| chr1_-_158686700 | 0.09 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1 |
| chr19_-_6670151 | 0.09 |
ENST00000675206.1
|
TNFSF14
|
TNF superfamily member 14 |
| chr22_-_19479160 | 0.09 |
ENST00000399523.5
ENST00000421968.6 ENST00000263202.15 ENST00000447868.5 |
UFD1
|
ubiquitin recognition factor in ER associated degradation 1 |
| chr7_-_19773569 | 0.09 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr1_-_150235943 | 0.09 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr2_-_61537740 | 0.09 |
ENST00000678081.1
ENST00000676889.1 ENST00000677850.1 ENST00000676789.1 |
XPO1
|
exportin 1 |
| chr15_+_59611776 | 0.09 |
ENST00000396065.3
ENST00000560585.5 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr17_-_67366562 | 0.09 |
ENST00000356126.8
ENST00000357146.4 |
PSMD12
|
proteasome 26S subunit, non-ATPase 12 |
| chrX_-_71106728 | 0.09 |
ENST00000374251.6
|
CXorf65
|
chromosome X open reading frame 65 |
| chr17_-_15263162 | 0.09 |
ENST00000674673.1
ENST00000675950.1 |
PMP22
|
peripheral myelin protein 22 |
| chr8_-_93740718 | 0.09 |
ENST00000519109.6
|
RBM12B
|
RNA binding motif protein 12B |
| chr3_+_69866217 | 0.09 |
ENST00000314589.10
|
MITF
|
melanocyte inducing transcription factor |
| chr20_+_10218808 | 0.09 |
ENST00000254976.7
ENST00000304886.6 |
SNAP25
|
synaptosome associated protein 25 |
| chr22_+_29306582 | 0.09 |
ENST00000616432.4
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest specific 2 like 1 |
| chr10_+_84139491 | 0.08 |
ENST00000372134.6
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr12_-_56636318 | 0.08 |
ENST00000549506.5
ENST00000379441.7 ENST00000551812.5 |
BAZ2A
|
bromodomain adjacent to zinc finger domain 2A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.6 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 0.4 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.3 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.1 | 0.2 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.7 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.1 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1903004 | flavin adenine dinucleotide metabolic process(GO:0072387) regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.0 | 0.0 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.1 | GO:0072277 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.9 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.6 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.6 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 1.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.0 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0061635 | negative regulation of growth of symbiont in host(GO:0044130) regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.3 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.7 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 1.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.4 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 3.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |