Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
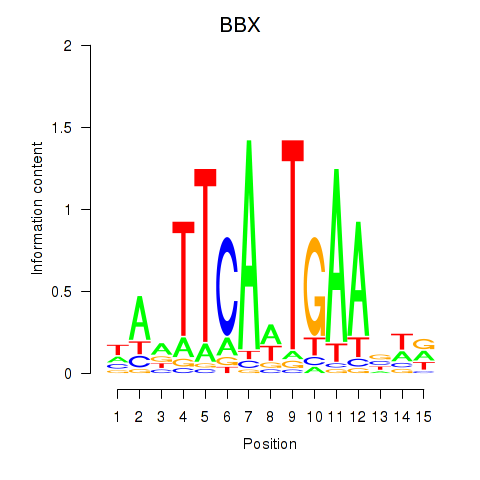
Results for BBX
Z-value: 0.35
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.19 | BBX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BBX | hg38_v1_chr3_+_107522936_107523003 | 0.31 | 9.9e-02 | Click! |
Activity profile of BBX motif
Sorted Z-values of BBX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BBX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_16881967 | 2.08 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr11_+_60699605 | 1.79 |
ENST00000300226.7
|
MS4A8
|
membrane spanning 4-domains A8 |
| chr1_-_161367872 | 1.36 |
ENST00000367974.2
|
CFAP126
|
cilia and flagella associated protein 126 |
| chr4_-_122621011 | 1.08 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr6_+_52420107 | 0.99 |
ENST00000636489.1
ENST00000637089.1 ENST00000637353.1 ENST00000637263.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr20_-_7940444 | 0.96 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr1_+_36084079 | 0.73 |
ENST00000207457.8
|
TEKT2
|
tektin 2 |
| chr11_-_22625804 | 0.42 |
ENST00000327470.6
|
FANCF
|
FA complementation group F |
| chr1_-_216805367 | 0.42 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr2_-_61854119 | 0.41 |
ENST00000405894.3
|
FAM161A
|
FAM161 centrosomal protein A |
| chr2_-_61854013 | 0.40 |
ENST00000404929.6
|
FAM161A
|
FAM161 centrosomal protein A |
| chr8_-_28490220 | 0.38 |
ENST00000517673.5
ENST00000380254.7 ENST00000518734.5 ENST00000346498.6 |
FBXO16
|
F-box protein 16 |
| chr8_-_144605699 | 0.37 |
ENST00000377307.6
ENST00000276826.5 |
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr2_-_119366682 | 0.36 |
ENST00000409877.5
ENST00000409523.1 ENST00000409466.6 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr6_-_135497230 | 0.34 |
ENST00000681365.1
|
AHI1
|
Abelson helper integration site 1 |
| chr13_+_76948500 | 0.32 |
ENST00000377462.6
|
ACOD1
|
aconitate decarboxylase 1 |
| chr12_-_100262356 | 0.31 |
ENST00000548313.5
|
DEPDC4
|
DEP domain containing 4 |
| chr6_+_31927486 | 0.30 |
ENST00000442278.6
|
C2
|
complement C2 |
| chr1_+_78620722 | 0.27 |
ENST00000679848.1
|
IFI44L
|
interferon induced protein 44 like |
| chr6_-_25874212 | 0.26 |
ENST00000361703.10
ENST00000397060.8 |
SLC17A3
|
solute carrier family 17 member 3 |
| chr17_+_21126947 | 0.25 |
ENST00000579303.5
|
DHRS7B
|
dehydrogenase/reductase 7B |
| chr12_-_39340963 | 0.25 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A |
| chr17_+_42798881 | 0.22 |
ENST00000588527.5
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr17_+_42798857 | 0.22 |
ENST00000588408.6
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr3_-_112499457 | 0.21 |
ENST00000334529.10
|
BTLA
|
B and T lymphocyte associated |
| chr3_-_112499358 | 0.21 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr1_+_158461574 | 0.21 |
ENST00000641432.1
ENST00000641460.1 ENST00000641535.1 ENST00000641971.1 |
OR10K1
|
olfactory receptor family 10 subfamily K member 1 |
| chr11_+_58622659 | 0.20 |
ENST00000361987.6
|
CNTF
|
ciliary neurotrophic factor |
| chr4_-_103099811 | 0.19 |
ENST00000504285.5
ENST00000296424.9 |
BDH2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr9_-_13175824 | 0.18 |
ENST00000545857.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr16_-_28363508 | 0.17 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family member B6 |
| chr12_+_55362975 | 0.17 |
ENST00000641576.1
|
OR6C75
|
olfactory receptor family 6 subfamily C member 75 |
| chr4_+_107824555 | 0.16 |
ENST00000394684.8
|
SGMS2
|
sphingomyelin synthase 2 |
| chr16_+_56191476 | 0.15 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr1_-_37514743 | 0.15 |
ENST00000448519.2
ENST00000373075.6 ENST00000373073.8 ENST00000296214.10 |
MEAF6
|
MYST/Esa1 associated factor 6 |
| chr10_-_92243246 | 0.15 |
ENST00000412050.8
ENST00000614585.4 |
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr3_-_146528750 | 0.15 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr14_+_63761836 | 0.15 |
ENST00000674003.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr5_+_126423122 | 0.15 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr6_+_117482634 | 0.15 |
ENST00000296955.12
ENST00000338728.10 |
DCBLD1
|
discoidin, CUB and LCCL domain containing 1 |
| chr21_+_30396030 | 0.15 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr14_+_67189393 | 0.14 |
ENST00000524532.5
ENST00000612183.4 ENST00000530728.5 |
FAM71D
|
family with sequence similarity 71 member D |
| chr17_+_68259164 | 0.14 |
ENST00000448504.6
|
ARSG
|
arylsulfatase G |
| chr1_-_153150884 | 0.13 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr5_+_162067500 | 0.13 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr4_-_158723355 | 0.13 |
ENST00000307720.4
|
PPID
|
peptidylprolyl isomerase D |
| chr6_-_49744434 | 0.13 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr16_+_28751787 | 0.12 |
ENST00000357796.7
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family member B9 |
| chr20_-_31722854 | 0.12 |
ENST00000307677.5
|
BCL2L1
|
BCL2 like 1 |
| chr19_+_6887560 | 0.12 |
ENST00000250572.12
ENST00000381407.9 ENST00000450315.7 ENST00000312053.9 ENST00000381404.8 |
ADGRE1
|
adhesion G protein-coupled receptor E1 |
| chr6_-_49744378 | 0.11 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr7_-_16833411 | 0.11 |
ENST00000412973.1
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr5_+_141135199 | 0.11 |
ENST00000231134.8
ENST00000623915.1 |
PCDHB5
|
protocadherin beta 5 |
| chr10_+_110005804 | 0.11 |
ENST00000360162.7
|
ADD3
|
adducin 3 |
| chr20_-_31722949 | 0.10 |
ENST00000376055.9
|
BCL2L1
|
BCL2 like 1 |
| chr6_-_29375291 | 0.10 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor family 12 subfamily D member 3 |
| chr9_+_35161951 | 0.10 |
ENST00000617908.4
ENST00000619578.4 |
UNC13B
|
unc-13 homolog B |
| chr21_+_18244828 | 0.10 |
ENST00000299295.7
|
CHODL
|
chondrolectin |
| chr9_-_5833014 | 0.09 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr4_-_185775890 | 0.09 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_58666765 | 0.09 |
ENST00000358781.7
|
FAM3D
|
FAM3 metabolism regulating signaling molecule D |
| chr19_+_17719471 | 0.09 |
ENST00000600186.5
ENST00000597735.5 ENST00000324096.9 |
MAP1S
|
microtubule associated protein 1S |
| chr4_-_112636858 | 0.08 |
ENST00000503172.5
ENST00000505019.6 ENST00000309071.9 |
ZGRF1
|
zinc finger GRF-type containing 1 |
| chr19_-_18397596 | 0.08 |
ENST00000595840.1
ENST00000339007.4 |
LRRC25
|
leucine rich repeat containing 25 |
| chrX_-_19799751 | 0.07 |
ENST00000379698.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr7_+_117480011 | 0.07 |
ENST00000649406.1
ENST00000648260.1 ENST00000003084.11 |
CFTR
|
CF transmembrane conductance regulator |
| chr20_-_31722533 | 0.07 |
ENST00000677194.1
ENST00000434194.2 ENST00000376062.6 |
BCL2L1
|
BCL2 like 1 |
| chr18_-_23437927 | 0.07 |
ENST00000578520.5
ENST00000383233.8 |
TMEM241
|
transmembrane protein 241 |
| chr9_+_35162000 | 0.07 |
ENST00000396787.5
ENST00000378495.7 ENST00000635942.1 ENST00000378496.8 |
UNC13B
|
unc-13 homolog B |
| chr15_-_75455767 | 0.07 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr17_-_75785486 | 0.07 |
ENST00000586607.5
|
H3-3B
|
H3.3 histone B |
| chr4_+_158315309 | 0.07 |
ENST00000460056.6
|
RXFP1
|
relaxin family peptide receptor 1 |
| chr5_+_162067458 | 0.07 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_-_62403506 | 0.06 |
ENST00000680062.1
|
DIMT1
|
DIMT1 rRNA methyltransferase and ribosome maturation factor |
| chr14_+_74763308 | 0.06 |
ENST00000325680.12
ENST00000552421.5 |
YLPM1
|
YLP motif containing 1 |
| chr8_-_107498041 | 0.06 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1 |
| chr4_-_185775271 | 0.06 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_+_72118392 | 0.06 |
ENST00000340912.6
|
SENP8
|
SUMO peptidase family member, NEDD8 specific |
| chr5_+_150661243 | 0.06 |
ENST00000517768.6
|
MYOZ3
|
myozenin 3 |
| chr15_-_78299570 | 0.05 |
ENST00000560569.5
ENST00000558459.5 ENST00000558311.5 ENST00000267973.7 |
WDR61
|
WD repeat domain 61 |
| chr8_-_107497909 | 0.05 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1 |
| chr9_-_2844058 | 0.05 |
ENST00000397885.3
|
PUM3
|
pumilio RNA binding family member 3 |
| chr3_+_148865288 | 0.05 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3 |
| chr9_-_92404559 | 0.05 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr9_+_101185029 | 0.05 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr14_+_22304051 | 0.05 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr3_+_122680802 | 0.05 |
ENST00000474629.7
|
PARP14
|
poly(ADP-ribose) polymerase family member 14 |
| chr14_+_20110739 | 0.05 |
ENST00000641386.2
ENST00000641633.2 |
OR4K17
|
olfactory receptor family 4 subfamily K member 17 |
| chr3_+_51817596 | 0.05 |
ENST00000456080.5
|
ENSG00000285749.2
|
novel protein |
| chr12_-_16600703 | 0.04 |
ENST00000616247.4
|
LMO3
|
LIM domain only 3 |
| chr3_-_169147734 | 0.04 |
ENST00000464456.5
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr6_+_54308429 | 0.04 |
ENST00000259782.9
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr5_+_141430565 | 0.04 |
ENST00000613314.1
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr5_-_59216826 | 0.04 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D |
| chr15_+_40351026 | 0.04 |
ENST00000448599.2
|
PHGR1
|
proline, histidine and glycine rich 1 |
| chr2_+_33436304 | 0.03 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr2_-_157325808 | 0.03 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr4_+_70592253 | 0.03 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr5_+_141182369 | 0.03 |
ENST00000609684.3
ENST00000625044.1 ENST00000623407.1 ENST00000623884.1 |
PCDHB16
ENSG00000279068.1
|
protocadherin beta 16 novel transcript |
| chr1_+_51617079 | 0.03 |
ENST00000447887.5
ENST00000428468.6 ENST00000453295.5 |
OSBPL9
|
oxysterol binding protein like 9 |
| chr2_+_112181788 | 0.02 |
ENST00000272559.4
|
FBLN7
|
fibulin 7 |
| chr17_+_29941605 | 0.02 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr3_+_111542178 | 0.02 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr2_-_230219902 | 0.02 |
ENST00000409815.6
|
SP110
|
SP110 nuclear body protein |
| chr5_+_150660841 | 0.02 |
ENST00000297130.4
|
MYOZ3
|
myozenin 3 |
| chr5_-_22853320 | 0.02 |
ENST00000504376.6
ENST00000382254.6 |
CDH12
|
cadherin 12 |
| chr4_-_993376 | 0.02 |
ENST00000398520.6
ENST00000398516.3 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr10_+_24208774 | 0.02 |
ENST00000376456.8
ENST00000458595.5 ENST00000376452.7 ENST00000430453.6 |
KIAA1217
|
KIAA1217 |
| chr1_-_13285154 | 0.01 |
ENST00000357367.6
ENST00000614831.1 |
PRAMEF8
|
PRAME family member 8 |
| chrX_-_126166273 | 0.01 |
ENST00000360028.4
|
DCAF12L2
|
DDB1 and CUL4 associated factor 12 like 2 |
| chr14_+_21868822 | 0.01 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr6_-_130222813 | 0.01 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr22_+_29438583 | 0.00 |
ENST00000354373.2
|
RFPL1
|
ret finger protein like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.3 | GO:0039008 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.1 | 1.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.3 | GO:0072573 | propionate metabolic process(GO:0019541) cellular response to progesterone stimulus(GO:0071393) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.0 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.2 | 1.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |