Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
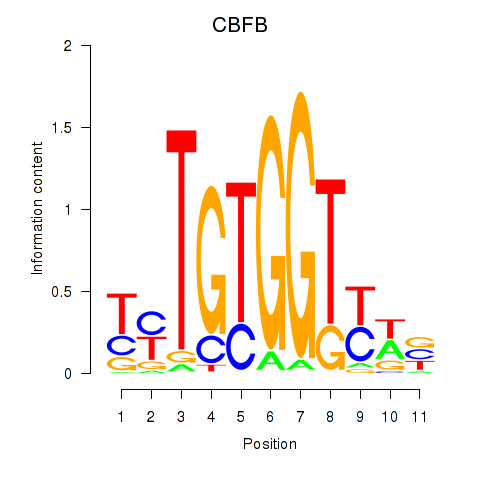
Results for CBFB
Z-value: 0.36
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.15 | CBFB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg38_v1_chr16_+_67029093_67029116 | 0.26 | 1.6e-01 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_85530127 | 0.82 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr20_+_46008900 | 0.74 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr22_+_31082860 | 0.61 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chr2_-_157444044 | 0.52 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr5_-_39202991 | 0.50 |
ENST00000515010.5
|
FYB1
|
FYN binding protein 1 |
| chr10_+_17229267 | 0.50 |
ENST00000224237.9
|
VIM
|
vimentin |
| chr11_-_58575846 | 0.48 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr14_+_85530163 | 0.48 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr2_-_127220293 | 0.41 |
ENST00000664447.2
ENST00000409327.2 |
CYP27C1
|
cytochrome P450 family 27 subfamily C member 1 |
| chr9_-_120876356 | 0.40 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr4_+_87975667 | 0.39 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr4_+_87975829 | 0.38 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr7_+_120988683 | 0.36 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr20_+_56412112 | 0.34 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4 |
| chr3_-_99850976 | 0.33 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like |
| chr3_+_98763331 | 0.32 |
ENST00000485391.5
ENST00000492254.5 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr20_+_56412249 | 0.32 |
ENST00000679887.1
ENST00000434344.2 |
CASS4
|
Cas scaffold protein family member 4 |
| chr20_+_56412393 | 0.31 |
ENST00000679529.1
|
CASS4
|
Cas scaffold protein family member 4 |
| chr12_-_52192007 | 0.30 |
ENST00000394815.3
|
KRT80
|
keratin 80 |
| chr12_-_52191981 | 0.29 |
ENST00000313234.9
|
KRT80
|
keratin 80 |
| chr6_+_26500296 | 0.29 |
ENST00000684113.1
|
BTN1A1
|
butyrophilin subfamily 1 member A1 |
| chr3_+_133124807 | 0.28 |
ENST00000508711.5
|
TMEM108
|
transmembrane protein 108 |
| chr3_+_9933805 | 0.21 |
ENST00000684493.1
ENST00000673935.2 ENST00000684181.1 ENST00000683189.1 ENST00000383811.8 ENST00000452070.6 ENST00000682642.1 ENST00000684659.1 ENST00000491527.2 ENST00000326434.9 ENST00000682783.1 ENST00000683835.1 ENST00000682570.1 |
CRELD1
|
cysteine rich with EGF like domains 1 |
| chr12_-_14696571 | 0.19 |
ENST00000261170.5
|
GUCY2C
|
guanylate cyclase 2C |
| chr13_-_46182136 | 0.19 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr12_+_14973020 | 0.19 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H |
| chrX_+_68829009 | 0.19 |
ENST00000204961.5
|
EFNB1
|
ephrin B1 |
| chr19_-_13937991 | 0.16 |
ENST00000254320.7
ENST00000586075.1 |
PODNL1
|
podocan like 1 |
| chr19_-_13938371 | 0.16 |
ENST00000588872.3
ENST00000339560.10 |
PODNL1
|
podocan like 1 |
| chr13_-_28100556 | 0.15 |
ENST00000241453.12
|
FLT3
|
fms related receptor tyrosine kinase 3 |
| chr3_-_190122317 | 0.15 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chrX_-_49184789 | 0.14 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr5_+_132073782 | 0.14 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2 |
| chr9_-_35079923 | 0.14 |
ENST00000378643.8
ENST00000448890.1 |
FANCG
|
FA complementation group G |
| chr1_+_209686173 | 0.14 |
ENST00000615289.4
ENST00000367028.6 ENST00000261465.5 |
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr1_-_240612147 | 0.13 |
ENST00000318160.5
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr8_-_59119121 | 0.12 |
ENST00000361421.2
|
TOX
|
thymocyte selection associated high mobility group box |
| chr15_+_73683938 | 0.12 |
ENST00000567189.5
|
CD276
|
CD276 molecule |
| chrX_-_71618455 | 0.12 |
ENST00000373691.4
ENST00000373693.4 |
CXCR3
|
C-X-C motif chemokine receptor 3 |
| chr14_-_59484006 | 0.12 |
ENST00000481608.1
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr2_+_102418642 | 0.11 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr4_+_70226116 | 0.11 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr1_-_184037695 | 0.11 |
ENST00000361927.9
ENST00000649786.1 |
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr4_+_73481737 | 0.11 |
ENST00000226355.5
|
AFM
|
afamin |
| chr11_+_114439424 | 0.10 |
ENST00000544196.5
ENST00000265881.10 ENST00000539754.5 ENST00000539275.5 |
REXO2
|
RNA exonuclease 2 |
| chrX_-_139708190 | 0.10 |
ENST00000414978.5
ENST00000519895.5 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr2_+_68365274 | 0.10 |
ENST00000234313.8
|
PLEK
|
pleckstrin |
| chr18_+_13611910 | 0.09 |
ENST00000590308.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr12_+_80716906 | 0.09 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr5_+_35856883 | 0.09 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr14_-_59484317 | 0.08 |
ENST00000247194.9
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr11_+_114439515 | 0.08 |
ENST00000539119.5
|
REXO2
|
RNA exonuclease 2 |
| chr1_+_34176900 | 0.07 |
ENST00000488417.2
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chr6_-_75202792 | 0.07 |
ENST00000416123.6
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr8_-_133060347 | 0.06 |
ENST00000427060.6
|
SLA
|
Src like adaptor |
| chr14_+_22281097 | 0.06 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr12_-_54634880 | 0.06 |
ENST00000547511.5
ENST00000257867.5 |
LACRT
|
lacritin |
| chr17_-_48101087 | 0.06 |
ENST00000393408.7
|
CBX1
|
chromobox 1 |
| chr17_-_48101379 | 0.05 |
ENST00000581003.5
ENST00000225603.9 |
CBX1
|
chromobox 1 |
| chr12_-_54384687 | 0.05 |
ENST00000550120.1
ENST00000547210.5 ENST00000394313.7 |
ZNF385A
|
zinc finger protein 385A |
| chr18_-_28177934 | 0.05 |
ENST00000676445.1
|
CDH2
|
cadherin 2 |
| chr17_+_34255274 | 0.04 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr17_+_66302606 | 0.04 |
ENST00000413366.8
|
PRKCA
|
protein kinase C alpha |
| chr15_+_64094060 | 0.03 |
ENST00000560829.5
|
SNX1
|
sorting nexin 1 |
| chr11_-_66336396 | 0.03 |
ENST00000627248.1
ENST00000311320.9 |
RIN1
|
Ras and Rab interactor 1 |
| chr3_+_157436842 | 0.03 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr13_+_75636311 | 0.03 |
ENST00000377499.9
ENST00000377534.8 |
LMO7
|
LIM domain 7 |
| chr5_-_88785493 | 0.03 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_-_68159732 | 0.02 |
ENST00000229135.4
|
IFNG
|
interferon gamma |
| chr6_+_27957241 | 0.02 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor family 2 subfamily B member 6 |
| chr1_-_184754808 | 0.01 |
ENST00000318130.13
ENST00000367512.7 |
EDEM3
|
ER degradation enhancing alpha-mannosidase like protein 3 |
| chr14_+_71598229 | 0.01 |
ENST00000537413.5
|
SIPA1L1
|
signal induced proliferation associated 1 like 1 |
| chr15_+_49423233 | 0.01 |
ENST00000560270.1
ENST00000267843.9 ENST00000560979.1 |
FGF7
|
fibroblast growth factor 7 |
| chr17_-_41027198 | 0.01 |
ENST00000361883.6
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr16_+_85611401 | 0.00 |
ENST00000405402.6
|
GSE1
|
Gse1 coiled-coil protein |
| chr9_+_15422704 | 0.00 |
ENST00000380821.7
ENST00000610884.4 ENST00000421710.5 |
SNAPC3
|
small nuclear RNA activating complex polypeptide 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 1.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.7 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.5 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.0 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.2 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |