Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for CDC5L
Z-value: 0.71
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.8 | CDC5L |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg38_v1_chr6_+_44387686_44387730 | 0.11 | 5.8e-01 | Click! |
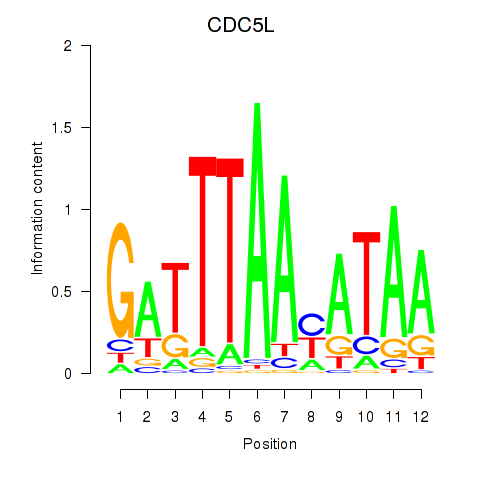
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_16881967 | 5.74 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr6_-_32589833 | 4.06 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr4_-_99352730 | 3.33 |
ENST00000510055.5
ENST00000515683.6 ENST00000511397.3 |
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide |
| chr6_-_32530268 | 2.60 |
ENST00000374975.4
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr1_+_37556913 | 2.37 |
ENST00000296218.8
ENST00000652629.1 |
DNALI1
|
dynein axonemal light intermediate chain 1 |
| chr5_-_35938572 | 2.19 |
ENST00000651391.1
ENST00000397366.5 ENST00000513623.5 ENST00000514524.2 ENST00000397367.6 |
CAPSL
|
calcyphosine like |
| chr17_+_47831608 | 1.91 |
ENST00000269025.9
|
LRRC46
|
leucine rich repeat containing 46 |
| chr5_+_140786136 | 1.83 |
ENST00000378133.4
ENST00000504120.4 |
PCDHA1
|
protocadherin alpha 1 |
| chr17_-_4786433 | 1.78 |
ENST00000354194.4
|
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr17_-_4786354 | 1.70 |
ENST00000328739.6
ENST00000441199.2 ENST00000416307.6 |
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr6_+_131637296 | 1.65 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr8_-_109680812 | 1.61 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr10_-_67838173 | 1.52 |
ENST00000225171.7
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr7_+_92057602 | 1.52 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr5_+_140834230 | 1.44 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr12_+_20810698 | 1.38 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr2_+_102104563 | 1.38 |
ENST00000409589.5
ENST00000409329.5 |
IL1R1
|
interleukin 1 receptor type 1 |
| chr3_-_120647018 | 1.32 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr15_+_70936487 | 1.26 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr6_-_165309598 | 1.20 |
ENST00000230301.9
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr8_-_132625378 | 1.16 |
ENST00000522789.5
|
LRRC6
|
leucine rich repeat containing 6 |
| chr10_-_46046264 | 1.10 |
ENST00000581478.5
ENST00000582163.3 |
MSMB
|
microseminoprotein beta |
| chr3_+_108602776 | 1.07 |
ENST00000497905.5
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr6_+_32844789 | 1.05 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr4_-_99435396 | 1.04 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_+_72192126 | 1.01 |
ENST00000393676.5
|
FOLR1
|
folate receptor alpha |
| chr8_+_75539862 | 1.00 |
ENST00000396423.4
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr1_-_150765735 | 0.99 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr1_+_47023659 | 0.98 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1 |
| chr6_-_52840843 | 0.97 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr4_-_99435134 | 0.95 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_-_112073980 | 0.95 |
ENST00000532211.5
ENST00000528775.6 ENST00000280350.10 ENST00000431456.6 |
PIH1D2
|
PIH1 domain containing 2 |
| chr1_+_103571077 | 0.94 |
ENST00000610648.1
|
AMY2B
|
amylase alpha 2B |
| chr14_-_105987068 | 0.94 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr8_+_75539893 | 0.94 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr19_-_8896090 | 0.93 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr4_-_149815826 | 0.92 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr4_-_38804783 | 0.91 |
ENST00000308979.7
ENST00000505940.1 ENST00000515861.5 |
TLR1
|
toll like receptor 1 |
| chr4_-_176195563 | 0.91 |
ENST00000280191.7
|
SPATA4
|
spermatogenesis associated 4 |
| chr3_-_167653952 | 0.90 |
ENST00000466760.5
ENST00000479765.5 |
WDR49
|
WD repeat domain 49 |
| chr3_+_93980203 | 0.90 |
ENST00000679607.1
ENST00000679587.1 |
ARL13B
|
ADP ribosylation factor like GTPase 13B |
| chr11_+_73950985 | 0.85 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr7_+_117014881 | 0.84 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr5_+_141135199 | 0.84 |
ENST00000231134.8
ENST00000623915.1 |
PCDHB5
|
protocadherin beta 5 |
| chr5_+_140848360 | 0.82 |
ENST00000532602.2
|
PCDHA9
|
protocadherin alpha 9 |
| chr11_-_112074239 | 0.82 |
ENST00000530641.5
|
PIH1D2
|
PIH1 domain containing 2 |
| chr10_-_114526804 | 0.80 |
ENST00000369266.7
ENST00000369253.6 |
ABLIM1
|
actin binding LIM protein 1 |
| chr4_+_41612892 | 0.80 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_-_20382461 | 0.79 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr12_+_133181409 | 0.77 |
ENST00000416488.5
ENST00000228289.9 ENST00000541211.6 ENST00000536435.7 ENST00000500625.7 ENST00000539248.6 ENST00000542711.6 ENST00000536899.6 ENST00000542986.6 ENST00000611984.4 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr18_+_58341038 | 0.75 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr9_+_2159672 | 0.74 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_85578345 | 0.74 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr4_-_109801978 | 0.73 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr19_-_11435098 | 0.73 |
ENST00000356392.9
ENST00000591179.5 |
ODAD3
|
outer dynein arm docking complex subunit 3 |
| chr2_+_69013170 | 0.71 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr12_+_133181529 | 0.71 |
ENST00000541009.6
ENST00000592241.5 |
ZNF268
|
zinc finger protein 268 |
| chr7_-_120858303 | 0.70 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr10_-_114526897 | 0.66 |
ENST00000428430.1
ENST00000392952.7 |
ABLIM1
|
actin binding LIM protein 1 |
| chr12_+_9827517 | 0.66 |
ENST00000537723.5
|
KLRF1
|
killer cell lectin like receptor F1 |
| chr3_-_167653916 | 0.65 |
ENST00000488012.5
ENST00000682715.1 ENST00000647816.1 |
WDR49
|
WD repeat domain 49 |
| chr2_-_221572272 | 0.65 |
ENST00000409854.5
ENST00000443796.5 ENST00000281821.7 |
EPHA4
|
EPH receptor A4 |
| chr1_-_86383078 | 0.64 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like |
| chr12_-_39340963 | 0.63 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A |
| chr2_-_174847765 | 0.63 |
ENST00000443238.6
|
CHN1
|
chimerin 1 |
| chr1_+_156369202 | 0.63 |
ENST00000537040.6
|
RHBG
|
Rh family B glycoprotein |
| chr4_+_94974984 | 0.63 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr14_-_21048431 | 0.62 |
ENST00000555026.5
|
NDRG2
|
NDRG family member 2 |
| chr10_-_114684457 | 0.62 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr10_-_114684612 | 0.62 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr10_-_60733467 | 0.61 |
ENST00000373827.6
|
ANK3
|
ankyrin 3 |
| chr11_+_36375978 | 0.60 |
ENST00000378867.7
|
PRR5L
|
proline rich 5 like |
| chr4_+_70242583 | 0.60 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr5_+_140786291 | 0.60 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1 |
| chr19_+_15949008 | 0.60 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr5_-_180072086 | 0.60 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr4_+_41612702 | 0.57 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_+_69565122 | 0.57 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr22_+_28772664 | 0.56 |
ENST00000448492.6
ENST00000421503.6 ENST00000249064.9 ENST00000444523.1 |
CCDC117
|
coiled-coil domain containing 117 |
| chr12_-_110920568 | 0.55 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr1_+_59310071 | 0.55 |
ENST00000371212.5
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr5_-_97142579 | 0.55 |
ENST00000274382.9
|
LIX1
|
limb and CNS expressed 1 |
| chr14_+_75985747 | 0.55 |
ENST00000679083.1
ENST00000314067.11 ENST00000238628.10 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 |
| chr6_-_49713564 | 0.54 |
ENST00000616725.4
ENST00000618917.4 |
CRISP2
|
cysteine rich secretory protein 2 |
| chr17_+_39700046 | 0.54 |
ENST00000269571.10
|
ERBB2
|
erb-b2 receptor tyrosine kinase 2 |
| chr6_-_75363003 | 0.53 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr11_-_85719111 | 0.53 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr17_+_70075215 | 0.52 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr15_-_55365231 | 0.52 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr5_+_140125935 | 0.52 |
ENST00000333305.5
|
IGIP
|
IgA inducing protein |
| chr7_-_120858066 | 0.51 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr12_+_12070932 | 0.51 |
ENST00000308721.9
|
BCL2L14
|
BCL2 like 14 |
| chr9_-_112890868 | 0.49 |
ENST00000374228.5
|
SLC46A2
|
solute carrier family 46 member 2 |
| chr7_+_116953379 | 0.48 |
ENST00000393449.5
|
ST7
|
suppression of tumorigenicity 7 |
| chr17_+_39700000 | 0.48 |
ENST00000584450.5
|
ERBB2
|
erb-b2 receptor tyrosine kinase 2 |
| chr4_-_23890035 | 0.46 |
ENST00000507380.1
ENST00000264867.7 |
PPARGC1A
|
PPARG coactivator 1 alpha |
| chr8_+_22053543 | 0.45 |
ENST00000519850.5
ENST00000381470.7 |
DMTN
|
dematin actin binding protein |
| chr11_-_1608463 | 0.45 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr11_-_85719045 | 0.45 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr10_-_67838019 | 0.44 |
ENST00000483798.6
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr16_-_46748337 | 0.44 |
ENST00000394809.9
|
MYLK3
|
myosin light chain kinase 3 |
| chr1_+_159015665 | 0.43 |
ENST00000567661.5
ENST00000474473.1 |
IFI16
|
interferon gamma inducible protein 16 |
| chr16_+_4795357 | 0.43 |
ENST00000586005.6
|
SMIM22
|
small integral membrane protein 22 |
| chr6_-_49713521 | 0.43 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2 |
| chr10_+_133394094 | 0.42 |
ENST00000477902.6
|
MTG1
|
mitochondrial ribosome associated GTPase 1 |
| chr9_-_123184233 | 0.42 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr1_-_89126066 | 0.42 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr3_+_171843337 | 0.41 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr13_-_41194485 | 0.41 |
ENST00000379483.4
|
KBTBD7
|
kelch repeat and BTB domain containing 7 |
| chr10_+_94938649 | 0.41 |
ENST00000461906.1
ENST00000260682.8 |
CYP2C9
|
cytochrome P450 family 2 subfamily C member 9 |
| chr4_+_41538143 | 0.41 |
ENST00000503057.6
ENST00000511496.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr13_+_102656933 | 0.41 |
ENST00000650757.1
|
TPP2
|
tripeptidyl peptidase 2 |
| chr5_-_111758061 | 0.41 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr6_-_28443463 | 0.40 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr17_+_39699960 | 0.39 |
ENST00000445658.6
|
ERBB2
|
erb-b2 receptor tyrosine kinase 2 |
| chr13_+_77741160 | 0.39 |
ENST00000314070.9
ENST00000351546.7 |
SLAIN1
|
SLAIN motif family member 1 |
| chr15_+_65530754 | 0.39 |
ENST00000566074.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr20_-_18497218 | 0.38 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase |
| chr10_+_94762673 | 0.37 |
ENST00000480405.2
ENST00000371321.9 |
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19 |
| chr2_-_110204967 | 0.37 |
ENST00000355301.8
ENST00000676053.1 ENST00000675067.1 ENST00000676028.1 ENST00000417665.5 ENST00000418527.1 ENST00000445609.7 ENST00000316534.8 ENST00000393272.7 |
NPHP1
|
nephrocystin 1 |
| chr10_-_122845850 | 0.37 |
ENST00000392790.6
|
CUZD1
|
CUB and zona pellucida like domains 1 |
| chr19_-_40090860 | 0.37 |
ENST00000599972.1
ENST00000450241.6 ENST00000595687.6 ENST00000340963.9 |
ZNF780A
|
zinc finger protein 780A |
| chr10_+_26438404 | 0.37 |
ENST00000356785.4
|
APBB1IP
|
amyloid beta precursor protein binding family B member 1 interacting protein |
| chr17_-_75779758 | 0.36 |
ENST00000592643.5
ENST00000591890.5 ENST00000587171.1 ENST00000254810.8 ENST00000589599.5 |
H3-3B
|
H3.3 histone B |
| chr15_+_65530418 | 0.36 |
ENST00000562901.5
ENST00000261875.10 ENST00000442729.6 ENST00000565299.5 ENST00000568793.5 |
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr12_+_54854505 | 0.36 |
ENST00000308796.11
ENST00000619042.1 |
MUCL1
|
mucin like 1 |
| chr5_+_140801028 | 0.36 |
ENST00000532566.3
ENST00000522353.3 |
PCDHA3
|
protocadherin alpha 3 |
| chrX_+_10158448 | 0.36 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr6_-_118710065 | 0.36 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr3_+_151814102 | 0.35 |
ENST00000232892.12
|
AADAC
|
arylacetamide deacetylase |
| chr17_-_37609361 | 0.35 |
ENST00000614941.4
ENST00000619541.4 ENST00000622045.4 ENST00000616179.4 ENST00000621136.4 ENST00000612223.5 ENST00000620424.1 |
SYNRG
|
synergin gamma |
| chr11_-_30992665 | 0.35 |
ENST00000406071.6
|
DCDC1
|
doublecortin domain containing 1 |
| chr9_-_38424446 | 0.35 |
ENST00000377694.2
|
IGFBPL1
|
insulin like growth factor binding protein like 1 |
| chr14_+_66824439 | 0.35 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr16_-_19886133 | 0.35 |
ENST00000568214.1
ENST00000569479.5 |
GPRC5B
|
G protein-coupled receptor class C group 5 member B |
| chr1_-_85404494 | 0.35 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr4_+_37453914 | 0.34 |
ENST00000381980.9
ENST00000508175.5 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr16_+_4795378 | 0.34 |
ENST00000588606.5
|
SMIM22
|
small integral membrane protein 22 |
| chr1_-_216805367 | 0.34 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr18_+_41955186 | 0.33 |
ENST00000639914.1
ENST00000262039.9 ENST00000398870.7 ENST00000586545.5 ENST00000585528.5 |
PIK3C3
|
phosphatidylinositol 3-kinase catalytic subunit type 3 |
| chr17_+_50834581 | 0.33 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr11_+_124185216 | 0.33 |
ENST00000318666.6
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr8_+_19313685 | 0.33 |
ENST00000265807.8
ENST00000518040.5 |
SH2D4A
|
SH2 domain containing 4A |
| chr3_+_190388120 | 0.33 |
ENST00000456423.2
ENST00000264734.3 |
CLDN16
|
claudin 16 |
| chr6_-_149648735 | 0.32 |
ENST00000335643.12
ENST00000444282.5 |
KATNA1
|
katanin catalytic subunit A1 |
| chr17_+_7884783 | 0.32 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr4_+_169620527 | 0.32 |
ENST00000360642.7
ENST00000512813.5 ENST00000513761.6 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr19_+_35448251 | 0.32 |
ENST00000599180.3
|
FFAR2
|
free fatty acid receptor 2 |
| chr5_+_140794832 | 0.32 |
ENST00000378132.2
ENST00000526136.2 ENST00000520672.2 |
PCDHA2
|
protocadherin alpha 2 |
| chr1_+_166989254 | 0.32 |
ENST00000367872.9
ENST00000447624.1 |
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr1_+_152663378 | 0.32 |
ENST00000368784.2
|
LCE2D
|
late cornified envelope 2D |
| chr5_-_134174765 | 0.32 |
ENST00000520417.1
|
SKP1
|
S-phase kinase associated protein 1 |
| chr1_+_196652022 | 0.31 |
ENST00000367429.9
ENST00000630130.2 ENST00000359637.2 |
CFH
|
complement factor H |
| chr12_-_64752871 | 0.31 |
ENST00000418919.6
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr6_-_139374605 | 0.31 |
ENST00000618718.1
ENST00000367651.4 |
CITED2
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 2 |
| chr2_+_168901290 | 0.31 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr15_-_55408245 | 0.31 |
ENST00000563171.5
ENST00000425574.7 ENST00000442196.8 ENST00000564092.1 |
CCPG1
|
cell cycle progression 1 |
| chrM_+_12329 | 0.31 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr2_-_24972032 | 0.31 |
ENST00000534855.5
|
DNAJC27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr15_-_43584504 | 0.31 |
ENST00000348806.10
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr1_+_166989089 | 0.31 |
ENST00000367870.6
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr11_+_6863057 | 0.30 |
ENST00000641461.1
|
OR10A2
|
olfactory receptor family 10 subfamily A member 2 |
| chr3_+_101724602 | 0.30 |
ENST00000341893.8
|
CEP97
|
centrosomal protein 97 |
| chr6_+_52186373 | 0.30 |
ENST00000648244.1
|
IL17A
|
interleukin 17A |
| chr6_+_52671080 | 0.30 |
ENST00000211314.5
|
TMEM14A
|
transmembrane protein 14A |
| chr2_+_178284907 | 0.30 |
ENST00000409631.5
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr16_+_57542672 | 0.30 |
ENST00000615867.4
ENST00000340339.4 |
ADGRG5
|
adhesion G protein-coupled receptor G5 |
| chr6_+_144583198 | 0.30 |
ENST00000367526.8
|
UTRN
|
utrophin |
| chr16_+_57542635 | 0.30 |
ENST00000349457.8
|
ADGRG5
|
adhesion G protein-coupled receptor G5 |
| chr11_-_107457801 | 0.29 |
ENST00000282251.9
|
CWF19L2
|
CWF19 like cell cycle control factor 2 |
| chr1_+_9539431 | 0.29 |
ENST00000302692.7
|
SLC25A33
|
solute carrier family 25 member 33 |
| chr3_-_197573323 | 0.29 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr4_-_84966637 | 0.29 |
ENST00000295888.9
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr5_-_157859096 | 0.29 |
ENST00000530742.5
ENST00000523908.5 ENST00000523094.5 ENST00000411809.7 |
CLINT1
|
clathrin interactor 1 |
| chr4_-_167234426 | 0.28 |
ENST00000541354.5
ENST00000509854.5 ENST00000512681.5 ENST00000357545.9 ENST00000510741.5 ENST00000510403.5 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr10_+_110005804 | 0.28 |
ENST00000360162.7
|
ADD3
|
adducin 3 |
| chr4_+_122339221 | 0.28 |
ENST00000442707.1
|
KIAA1109
|
KIAA1109 |
| chr11_-_31509588 | 0.28 |
ENST00000534812.5
ENST00000529749.5 ENST00000532287.6 ENST00000278200.5 ENST00000530023.5 ENST00000533642.1 |
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr1_-_33182030 | 0.27 |
ENST00000291416.10
|
TRIM62
|
tripartite motif containing 62 |
| chr4_-_167234579 | 0.27 |
ENST00000502330.5
ENST00000357154.7 ENST00000421836.6 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr2_+_231708511 | 0.27 |
ENST00000341369.11
ENST00000409115.8 ENST00000409683.5 |
PTMA
|
prothymosin alpha |
| chr12_-_55842950 | 0.27 |
ENST00000548629.5
|
MMP19
|
matrix metallopeptidase 19 |
| chr1_+_174799895 | 0.27 |
ENST00000489615.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr11_-_31509569 | 0.26 |
ENST00000526776.5
|
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr21_-_42395943 | 0.26 |
ENST00000398405.5
|
TMPRSS3
|
transmembrane serine protease 3 |
| chr3_-_94028582 | 0.26 |
ENST00000315099.3
|
STX19
|
syntaxin 19 |
| chr6_-_15586006 | 0.26 |
ENST00000462989.6
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr3_+_108822778 | 0.26 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr3_+_108822759 | 0.26 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr4_+_169620509 | 0.26 |
ENST00000347613.8
|
CLCN3
|
chloride voltage-gated channel 3 |
| chr11_-_236326 | 0.26 |
ENST00000525237.1
ENST00000382743.9 ENST00000532956.5 ENST00000525319.5 ENST00000524564.5 |
SIRT3
|
sirtuin 3 |
| chr10_-_27243019 | 0.26 |
ENST00000679293.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr11_+_103109522 | 0.25 |
ENST00000334267.11
|
DYNC2H1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr9_-_111330224 | 0.25 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr3_-_185552554 | 0.25 |
ENST00000424591.6
ENST00000296252.9 |
LIPH
|
lipase H |
| chr1_+_164559173 | 0.24 |
ENST00000420696.7
|
PBX1
|
PBX homeobox 1 |
| chr12_+_11171166 | 0.24 |
ENST00000622602.2
|
SMIM10L1
|
small integral membrane protein 10 like 1 |
| chr2_+_66435116 | 0.24 |
ENST00000272369.14
ENST00000560281.6 |
MEIS1
|
Meis homeobox 1 |
| chr14_+_19743571 | 0.24 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3 |
| chr1_+_151612001 | 0.23 |
ENST00000642376.1
ENST00000368843.8 ENST00000458013.7 |
SNX27
|
sorting nexin 27 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.6 | 1.7 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.5 | 2.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.5 | 1.5 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.5 | 1.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 1.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.3 | 1.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 0.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 3.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 0.7 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.2 | 0.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.6 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.9 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 1.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 0.5 | GO:2000182 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 0.6 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.4 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.1 | 0.3 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.3 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.1 | 0.8 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 1.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.3 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.6 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 1.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.2 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.9 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.0 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.3 | GO:0061428 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 1.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.8 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0036404 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 0.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 1.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.4 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0090164 | protein retention in Golgi apparatus(GO:0045053) asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 | 0.3 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 3.3 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.5 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 2.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.1 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 1.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.5 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.0 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 1.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.1 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 6.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.7 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 0.6 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 1.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.0 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 1.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 2.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0033167 | ARC complex(GO:0033167) |
| 0.0 | 1.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 1.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 1.7 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 0.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.0 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.2 | 0.7 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 0.5 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 5.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 2.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 6.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.3 | GO:0008940 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.7 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.2 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.7 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.3 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 6.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.4 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |