Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
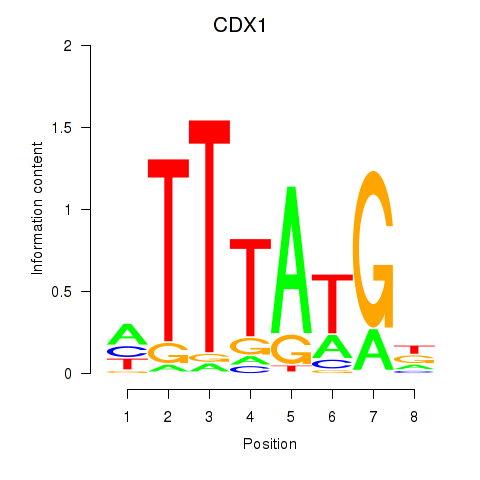
Results for CDX1
Z-value: 0.58
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.17 | CDX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX1 | hg38_v1_chr5_+_150166790_150166890, hg38_v1_chr5_+_150166770_150166788 | 0.20 | 2.8e-01 | Click! |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_68447453 | 1.90 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr3_+_190615308 | 1.77 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr11_-_102955705 | 1.76 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr1_+_20589044 | 1.63 |
ENST00000375071.4
|
CDA
|
cytidine deaminase |
| chr1_-_152414256 | 1.54 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr5_-_39202991 | 1.39 |
ENST00000515010.5
|
FYB1
|
FYN binding protein 1 |
| chr2_-_31138041 | 1.32 |
ENST00000324589.9
|
GALNT14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr12_-_52385649 | 1.32 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr10_-_88952763 | 1.16 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle |
| chr7_+_80638662 | 1.14 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chrX_+_100644183 | 1.07 |
ENST00000640889.1
ENST00000373004.5 |
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr17_-_41397600 | 1.02 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr6_+_126340107 | 0.96 |
ENST00000368328.5
ENST00000368326.5 ENST00000368325.5 |
CENPW
|
centromere protein W |
| chr1_+_84164962 | 0.95 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_-_149377637 | 0.91 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr16_-_18876305 | 0.89 |
ENST00000563235.5
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr2_-_207166818 | 0.87 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr13_+_77535742 | 0.87 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr2_-_207167220 | 0.84 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr20_+_6767678 | 0.83 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2 |
| chr12_-_27971970 | 0.79 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr2_+_33134620 | 0.78 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_84164370 | 0.77 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr17_-_59151794 | 0.77 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_+_33134579 | 0.75 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_47771029 | 0.73 |
ENST00000549151.5
ENST00000548919.5 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr9_-_92482350 | 0.73 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr3_+_172754457 | 0.73 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr8_-_100706931 | 0.71 |
ENST00000520868.5
|
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr9_+_74615582 | 0.68 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr17_-_43022350 | 0.66 |
ENST00000587173.5
ENST00000355653.8 |
VAT1
|
vesicle amine transport 1 |
| chr5_-_16742221 | 0.66 |
ENST00000505695.5
|
MYO10
|
myosin X |
| chr11_+_111245725 | 0.66 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr3_+_130931893 | 0.66 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr4_-_22443110 | 0.65 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr12_-_27972725 | 0.65 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr1_-_120054225 | 0.64 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr18_-_12656716 | 0.58 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr7_+_80638510 | 0.57 |
ENST00000433696.6
ENST00000538969.5 ENST00000544133.5 |
CD36
|
CD36 molecule |
| chr6_+_36678699 | 0.57 |
ENST00000405375.5
ENST00000244741.10 ENST00000373711.3 |
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr12_-_56934403 | 0.56 |
ENST00000293502.2
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C member 7 |
| chr6_+_31739948 | 0.56 |
ENST00000375755.8
ENST00000425703.5 ENST00000375750.9 ENST00000375703.7 ENST00000375740.7 |
MSH5
|
mutS homolog 5 |
| chr11_-_5516690 | 0.55 |
ENST00000380184.2
|
UBQLNL
|
ubiquilin like |
| chr9_-_21482313 | 0.55 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr7_+_80646436 | 0.54 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr13_+_52455972 | 0.54 |
ENST00000490903.5
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr3_+_101827982 | 0.53 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr7_+_80638633 | 0.53 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr3_-_151329539 | 0.53 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr1_+_40988513 | 0.53 |
ENST00000649215.1
|
CTPS1
|
CTP synthase 1 |
| chr2_+_10123171 | 0.53 |
ENST00000615152.5
|
RRM2
|
ribonucleotide reductase regulatory subunit M2 |
| chr8_+_22565236 | 0.52 |
ENST00000523900.5
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_+_178725147 | 0.52 |
ENST00000367634.6
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_-_120051714 | 0.51 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr18_+_63777773 | 0.50 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr9_-_92482499 | 0.49 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr11_+_117179127 | 0.48 |
ENST00000278951.11
|
SIDT2
|
SID1 transmembrane family member 2 |
| chr17_+_59155726 | 0.48 |
ENST00000578777.5
ENST00000577457.1 ENST00000582995.5 ENST00000262293.9 ENST00000614081.1 |
PRR11
|
proline rich 11 |
| chr5_+_36166556 | 0.48 |
ENST00000677886.1
|
SKP2
|
S-phase kinase associated protein 2 |
| chr3_-_193554799 | 0.48 |
ENST00000295548.3
|
ATP13A4
|
ATPase 13A4 |
| chr11_+_134069060 | 0.47 |
ENST00000534549.5
ENST00000441717.3 ENST00000299106.9 |
JAM3
|
junctional adhesion molecule 3 |
| chrX_+_108044967 | 0.47 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_197260369 | 0.45 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr15_+_41774539 | 0.45 |
ENST00000514566.5
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr11_-_11353241 | 0.45 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr17_+_44846318 | 0.44 |
ENST00000591513.5
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr1_+_153031195 | 0.44 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B |
| chr11_+_28108248 | 0.44 |
ENST00000406787.7
ENST00000403099.5 ENST00000407364.8 |
METTL15
|
methyltransferase like 15 |
| chr8_-_42207667 | 0.43 |
ENST00000352041.7
ENST00000679151.1 ENST00000679300.1 |
PLAT
|
plasminogen activator, tissue type |
| chrX_-_15664798 | 0.43 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator |
| chr3_-_197184131 | 0.43 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr14_-_67412112 | 0.42 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr12_+_12891554 | 0.41 |
ENST00000014914.6
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A |
| chr6_-_132763424 | 0.41 |
ENST00000532012.1
ENST00000525270.5 ENST00000530536.5 ENST00000524919.5 |
VNN2
|
vanin 2 |
| chr18_+_63775395 | 0.41 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr7_+_74773962 | 0.41 |
ENST00000289473.10
|
NCF1
|
neutrophil cytosolic factor 1 |
| chr8_-_42207557 | 0.40 |
ENST00000220809.9
ENST00000429089.6 ENST00000519510.5 ENST00000429710.6 ENST00000524009.5 |
PLAT
|
plasminogen activator, tissue type |
| chr18_+_63775369 | 0.40 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr10_+_100997040 | 0.40 |
ENST00000370223.7
|
LZTS2
|
leucine zipper tumor suppressor 2 |
| chr5_+_96743536 | 0.40 |
ENST00000515663.5
|
CAST
|
calpastatin |
| chr10_-_49134021 | 0.40 |
ENST00000311787.6
|
FAM170B
|
family with sequence similarity 170 member B |
| chr4_-_70666492 | 0.40 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr1_+_178341445 | 0.39 |
ENST00000462775.5
|
RASAL2
|
RAS protein activator like 2 |
| chr15_+_80441229 | 0.39 |
ENST00000533983.5
ENST00000527771.5 ENST00000525103.1 |
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr14_-_106639589 | 0.39 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr7_+_141764097 | 0.39 |
ENST00000247879.2
|
TAS2R3
|
taste 2 receptor member 3 |
| chr3_-_33645433 | 0.39 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_-_151227881 | 0.39 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr7_+_141790217 | 0.38 |
ENST00000247883.5
|
TAS2R5
|
taste 2 receptor member 5 |
| chr9_+_128340474 | 0.38 |
ENST00000300456.5
|
SLC27A4
|
solute carrier family 27 member 4 |
| chr6_-_47042260 | 0.37 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chrX_+_108045050 | 0.37 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr8_+_38728550 | 0.37 |
ENST00000520340.5
ENST00000518415.5 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr18_-_26863187 | 0.37 |
ENST00000440832.7
|
AQP4
|
aquaporin 4 |
| chr4_+_153152163 | 0.37 |
ENST00000676423.1
ENST00000675745.1 ENST00000676348.1 ENST00000676408.1 ENST00000674874.1 ENST00000675315.1 ENST00000675518.1 |
TRIM2
ENSG00000288637.1
|
tripartite motif containing 2 novel protein |
| chr8_-_37940103 | 0.37 |
ENST00000524298.1
ENST00000307599.5 |
GOT1L1
|
glutamic-oxaloacetic transaminase 1 like 1 |
| chr15_+_41774459 | 0.37 |
ENST00000457542.7
ENST00000456763.6 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr3_+_183265302 | 0.36 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr14_+_65411845 | 0.36 |
ENST00000556518.5
ENST00000557164.5 |
FUT8
|
fucosyltransferase 8 |
| chr16_+_56638659 | 0.35 |
ENST00000290705.12
|
MT1A
|
metallothionein 1A |
| chr10_+_5446601 | 0.34 |
ENST00000449083.5
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr6_-_116829037 | 0.34 |
ENST00000368549.7
ENST00000530250.1 ENST00000310357.8 |
GPRC6A
|
G protein-coupled receptor class C group 6 member A |
| chr15_+_71547226 | 0.33 |
ENST00000357769.4
ENST00000261862.7 |
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr5_-_128339191 | 0.33 |
ENST00000507835.5
|
FBN2
|
fibrillin 2 |
| chr16_-_56668034 | 0.33 |
ENST00000569500.5
ENST00000379811.4 ENST00000444837.6 |
MT1G
|
metallothionein 1G |
| chr3_+_150408314 | 0.32 |
ENST00000361875.7
|
TSC22D2
|
TSC22 domain family member 2 |
| chrX_+_12906639 | 0.32 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8 |
| chr17_-_10373002 | 0.31 |
ENST00000252172.9
ENST00000418404.8 |
MYH13
|
myosin heavy chain 13 |
| chr19_-_15418979 | 0.31 |
ENST00000397410.10
ENST00000596195.5 ENST00000595067.1 ENST00000595465.6 ENST00000680649.1 ENST00000679638.1 ENST00000600247.5 |
AKAP8L
|
A-kinase anchoring protein 8 like |
| chr3_+_159069252 | 0.31 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chrX_+_12906612 | 0.31 |
ENST00000218032.7
|
TLR8
|
toll like receptor 8 |
| chr2_-_162536955 | 0.30 |
ENST00000621889.1
|
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr13_+_31945826 | 0.30 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr3_-_37174578 | 0.30 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr7_-_97872394 | 0.29 |
ENST00000455086.5
ENST00000394308.8 ENST00000453600.5 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr14_-_23035223 | 0.29 |
ENST00000425762.2
|
PSMB5
|
proteasome 20S subunit beta 5 |
| chr7_+_144000320 | 0.29 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr12_+_96912517 | 0.29 |
ENST00000457368.2
|
NEDD1
|
NEDD1 gamma-tubulin ring complex targeting factor |
| chr7_-_97872120 | 0.28 |
ENST00000394309.7
ENST00000414884.1 ENST00000442734.5 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr12_-_10172117 | 0.28 |
ENST00000545927.5
ENST00000309539.8 ENST00000432556.6 ENST00000544577.5 |
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr7_+_77840122 | 0.28 |
ENST00000450574.5
ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr16_+_56657999 | 0.28 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr14_+_22040576 | 0.28 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr17_-_76570544 | 0.27 |
ENST00000640006.1
|
ENSG00000284526.1
|
novel protein |
| chr19_-_48993300 | 0.27 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr8_+_22565655 | 0.27 |
ENST00000523965.5
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr16_+_56657924 | 0.27 |
ENST00000334350.7
|
MT1F
|
metallothionein 1F |
| chr5_+_141421020 | 0.26 |
ENST00000622044.1
ENST00000398587.7 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr16_+_14833713 | 0.26 |
ENST00000287667.12
ENST00000620755.4 ENST00000610363.4 |
NOMO1
|
NODAL modulator 1 |
| chrX_+_41689006 | 0.26 |
ENST00000378138.5
ENST00000620846.1 ENST00000649219.1 |
GPR34
|
G protein-coupled receptor 34 |
| chr1_-_28058087 | 0.26 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr18_+_58149314 | 0.26 |
ENST00000435432.6
ENST00000357895.9 ENST00000586263.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr3_-_197226351 | 0.26 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr5_+_87267792 | 0.25 |
ENST00000274376.11
|
RASA1
|
RAS p21 protein activator 1 |
| chr15_+_51829644 | 0.25 |
ENST00000308580.12
|
TMOD3
|
tropomodulin 3 |
| chr3_+_136022734 | 0.25 |
ENST00000334546.6
|
PPP2R3A
|
protein phosphatase 2 regulatory subunit B''alpha |
| chrX_+_41688967 | 0.25 |
ENST00000378142.9
|
GPR34
|
G protein-coupled receptor 34 |
| chr14_+_88824621 | 0.25 |
ENST00000622513.4
ENST00000380656.7 ENST00000338104.10 ENST00000346301.8 ENST00000354441.10 ENST00000556651.5 ENST00000554686.5 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chrM_-_14669 | 0.25 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 |
| chr14_-_91732059 | 0.25 |
ENST00000553329.5
ENST00000256343.8 |
CATSPERB
|
cation channel sperm associated auxiliary subunit beta |
| chr14_+_22147988 | 0.25 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr7_+_107583919 | 0.25 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr2_+_135586250 | 0.25 |
ENST00000410054.5
ENST00000628915.2 |
R3HDM1
|
R3H domain containing 1 |
| chr7_+_120988683 | 0.25 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr5_+_80035341 | 0.24 |
ENST00000350881.6
|
THBS4
|
thrombospondin 4 |
| chr2_+_113005454 | 0.24 |
ENST00000259211.7
|
IL36A
|
interleukin 36 alpha |
| chr7_+_143959927 | 0.24 |
ENST00000624504.1
|
OR2F1
|
olfactory receptor family 2 subfamily F member 1 |
| chr14_+_96256194 | 0.24 |
ENST00000216629.11
ENST00000553356.1 |
BDKRB1
|
bradykinin receptor B1 |
| chr18_-_22417910 | 0.24 |
ENST00000391403.4
|
CTAGE1
|
cutaneous T cell lymphoma-associated antigen 1 |
| chr19_-_12957198 | 0.24 |
ENST00000316939.3
|
GADD45GIP1
|
GADD45G interacting protein 1 |
| chr7_+_106865474 | 0.24 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr8_+_2045058 | 0.24 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr3_-_197260722 | 0.24 |
ENST00000654733.1
ENST00000661808.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr1_+_159587817 | 0.24 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum |
| chr8_+_73991345 | 0.23 |
ENST00000284818.7
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr2_-_208129824 | 0.23 |
ENST00000282141.4
|
CRYGC
|
crystallin gamma C |
| chr8_-_143973520 | 0.23 |
ENST00000356346.7
|
PLEC
|
plectin |
| chr1_-_39883434 | 0.23 |
ENST00000541099.5
ENST00000441669.6 ENST00000316891.10 ENST00000537440.5 ENST00000372818.5 |
TRIT1
|
tRNA isopentenyltransferase 1 |
| chr4_-_82844418 | 0.23 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr10_-_32935511 | 0.23 |
ENST00000423113.5
|
ITGB1
|
integrin subunit beta 1 |
| chr6_+_26087417 | 0.23 |
ENST00000357618.10
ENST00000309234.10 |
HFE
|
homeostatic iron regulator |
| chr12_+_18262730 | 0.22 |
ENST00000675017.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr16_+_21678514 | 0.22 |
ENST00000286149.8
ENST00000388958.8 |
OTOA
|
otoancorin |
| chr15_-_82806054 | 0.22 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr9_+_74497308 | 0.22 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr14_+_21918161 | 0.22 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr11_-_13496018 | 0.22 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr2_+_161136901 | 0.22 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr6_-_127900958 | 0.22 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chr18_-_3845292 | 0.21 |
ENST00000400145.6
|
DLGAP1
|
DLG associated protein 1 |
| chr16_+_56651885 | 0.21 |
ENST00000334346.3
ENST00000562399.1 |
MT1B
|
metallothionein 1B |
| chr2_-_88128049 | 0.21 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chr2_+_172928165 | 0.21 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr17_-_58415628 | 0.20 |
ENST00000583753.5
|
RNF43
|
ring finger protein 43 |
| chr4_-_69961007 | 0.20 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr22_+_17628828 | 0.20 |
ENST00000399782.5
|
BCL2L13
|
BCL2 like 13 |
| chr9_+_122614738 | 0.20 |
ENST00000297913.3
|
OR1Q1
|
olfactory receptor family 1 subfamily Q member 1 |
| chr8_+_2045037 | 0.20 |
ENST00000262113.9
|
MYOM2
|
myomesin 2 |
| chr17_+_42844573 | 0.20 |
ENST00000253799.8
ENST00000452774.2 |
AOC2
|
amine oxidase copper containing 2 |
| chr13_-_33205997 | 0.20 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr3_+_44874606 | 0.20 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr20_+_16748358 | 0.20 |
ENST00000246081.3
|
OTOR
|
otoraplin |
| chr5_-_147831663 | 0.20 |
ENST00000296695.10
|
SPINK1
|
serine peptidase inhibitor Kazal type 1 |
| chr2_+_186694007 | 0.19 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr7_+_151028422 | 0.19 |
ENST00000542328.5
ENST00000461373.5 ENST00000297504.10 ENST00000358849.9 ENST00000498578.5 ENST00000477719.5 ENST00000477092.5 |
ABCB8
|
ATP binding cassette subfamily B member 8 |
| chr5_+_96743578 | 0.19 |
ENST00000325674.11
|
CAST
|
calpastatin |
| chr11_+_114439424 | 0.19 |
ENST00000544196.5
ENST00000265881.10 ENST00000539754.5 ENST00000539275.5 |
REXO2
|
RNA exonuclease 2 |
| chr8_-_673547 | 0.19 |
ENST00000522893.1
|
ERICH1
|
glutamate rich 1 |
| chr19_-_55407719 | 0.19 |
ENST00000587845.5
ENST00000589978.1 ENST00000264552.14 |
UBE2S
|
ubiquitin conjugating enzyme E2 S |
| chr13_+_45464901 | 0.19 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr6_+_29111560 | 0.19 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr10_-_67838091 | 0.19 |
ENST00000339758.7
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chrX_-_120559889 | 0.18 |
ENST00000371323.3
|
CUL4B
|
cullin 4B |
| chrX_-_139832235 | 0.18 |
ENST00000327569.7
ENST00000361648.6 |
ATP11C
|
ATPase phospholipid transporting 11C |
| chr2_-_150487658 | 0.18 |
ENST00000375734.6
ENST00000263895.9 ENST00000454202.5 |
RND3
|
Rho family GTPase 3 |
| chr2_+_169069537 | 0.18 |
ENST00000428522.5
ENST00000450153.1 ENST00000674881.1 ENST00000421653.5 |
DHRS9
|
dehydrogenase/reductase 9 |
| chr6_-_32407123 | 0.18 |
ENST00000374993.4
ENST00000544175.2 ENST00000454136.7 ENST00000446536.2 |
BTNL2
|
butyrophilin like 2 |
| chr9_-_96383675 | 0.18 |
ENST00000375257.2
ENST00000375259.9 ENST00000253270.13 |
SLC35D2
|
solute carrier family 35 member D2 |
| chr10_+_4963406 | 0.18 |
ENST00000380872.9
ENST00000442997.5 |
AKR1C1
|
aldo-keto reductase family 1 member C1 |
| chr15_-_66504832 | 0.18 |
ENST00000569438.2
ENST00000569696.5 ENST00000307961.11 |
RPL4
|
ribosomal protein L4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.3 | 2.8 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.3 | 0.8 | GO:0060128 | regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.3 | 1.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 1.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 1.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 1.2 | GO:0090131 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.2 | 1.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 1.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.6 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 0.4 | GO:1904440 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.6 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.4 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 0.4 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.5 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.8 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.4 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.8 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.1 | 1.8 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.3 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.2 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.4 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.2 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.7 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.6 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 1.3 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.2 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.6 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 1.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0010710 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.2 | GO:0010751 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1905006 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of alkaline phosphatase activity(GO:0010693) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.2 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.0 | 0.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.4 | GO:1903690 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 1.9 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.4 | GO:0071756 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.7 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 2.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 1.4 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 2.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 1.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 1.8 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 1.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.4 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.6 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.5 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 1.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0010465 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 2.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |