Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
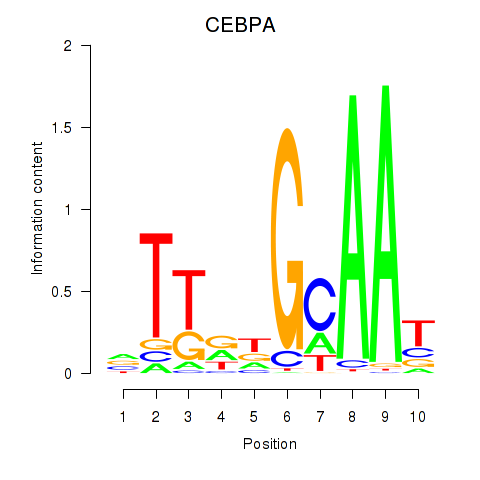
Results for CEBPA
Z-value: 1.14
Transcription factors associated with CEBPA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPA
|
ENSG00000245848.3 | CEBPA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPA | hg38_v1_chr19_-_33302524_33302542 | 0.22 | 2.5e-01 | Click! |
Activity profile of CEBPA motif
Sorted Z-values of CEBPA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.7 | 2.2 | GO:0010933 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.7 | 2.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.6 | 1.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 1.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.5 | 2.9 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.4 | 1.7 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.4 | 2.5 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.3 | 1.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 | 1.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.3 | 1.3 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 0.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 0.6 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.2 | 1.0 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 1.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 1.0 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 0.6 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.7 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.2 | 0.5 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 0.6 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.2 | 0.8 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.2 | 1.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.7 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.1 | 0.9 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.5 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 2.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.5 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 1.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.3 | GO:0002892 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.3 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 0.7 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.3 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 0.1 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 1.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 1.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.8 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 6.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.2 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.5 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.6 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.5 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.2 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.3 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.2 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.2 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.2 | GO:1902161 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 2.7 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.7 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.6 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 2.0 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.7 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.9 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0034201 | adiponectin-activated signaling pathway(GO:0033211) response to oleic acid(GO:0034201) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.4 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 1.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.2 | GO:0033152 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 1.7 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 1.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.3 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0032661 | chronic inflammatory response to antigenic stimulus(GO:0002439) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.5 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0060081 | oxalate transport(GO:0019532) membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:1903936 | cellular response to salt(GO:1902075) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.7 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:1990742 | microvesicle(GO:1990742) |
| 0.2 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.8 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 4.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 1.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 2.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 1.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 6.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 3.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 2.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.7 | 3.7 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.6 | 1.9 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.5 | 1.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 2.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 1.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.3 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 1.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.3 | 1.3 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 0.7 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 1.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 1.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.6 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 0.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.5 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 1.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 0.6 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.2 | 0.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.5 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 4.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.2 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.2 | GO:0004040 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 2.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.9 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 1.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 5.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.3 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 2.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.7 | GO:0023023 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 3.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 3.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 3.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 2.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |