Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
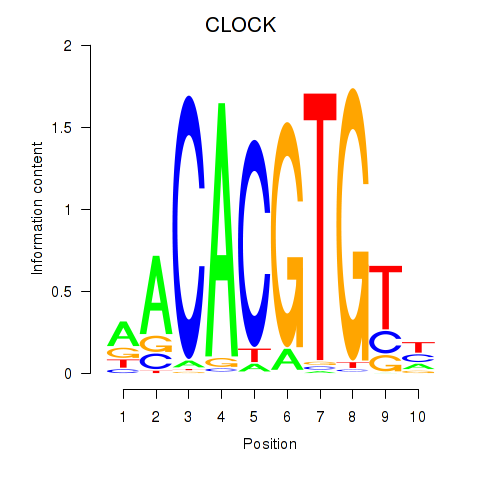
Results for CLOCK
Z-value: 0.66
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CLOCK
|
ENSG00000134852.15 | CLOCK |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CLOCK | hg38_v1_chr4_-_55546900_55546919 | -0.01 | 9.6e-01 | Click! |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_25906457 | 2.53 |
ENST00000426559.6
|
STMN1
|
stathmin 1 |
| chr18_+_36297661 | 1.37 |
ENST00000257209.8
ENST00000590592.5 ENST00000359247.8 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr20_+_45812576 | 1.11 |
ENST00000405520.5
ENST00000617055.4 |
UBE2C
|
ubiquitin conjugating enzyme E2 C |
| chr20_+_45812632 | 1.08 |
ENST00000335046.7
ENST00000356455.9 ENST00000243893.10 |
UBE2C
|
ubiquitin conjugating enzyme E2 C |
| chr1_-_25906411 | 1.07 |
ENST00000455785.7
|
STMN1
|
stathmin 1 |
| chr20_+_45812665 | 1.07 |
ENST00000352551.9
|
UBE2C
|
ubiquitin conjugating enzyme E2 C |
| chr14_-_58427134 | 1.06 |
ENST00000555930.6
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr6_+_69232406 | 1.03 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr2_-_215393126 | 0.99 |
ENST00000456923.5
|
FN1
|
fibronectin 1 |
| chr2_-_10448318 | 0.98 |
ENST00000234111.9
|
ODC1
|
ornithine decarboxylase 1 |
| chr8_-_94896660 | 0.98 |
ENST00000520509.5
|
CCNE2
|
cyclin E2 |
| chr11_+_6845683 | 0.94 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr2_-_10447771 | 0.93 |
ENST00000405333.5
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr5_+_136059151 | 0.89 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr1_-_11060000 | 0.89 |
ENST00000376957.7
|
SRM
|
spermidine synthase |
| chr9_-_120877026 | 0.88 |
ENST00000436309.5
|
PHF19
|
PHD finger protein 19 |
| chr9_-_132669969 | 0.80 |
ENST00000438527.7
|
DDX31
|
DEAD-box helicase 31 |
| chr20_-_51023081 | 0.80 |
ENST00000433903.5
ENST00000424171.5 ENST00000371571.5 ENST00000439216.5 |
KCNG1
|
potassium voltage-gated channel modifier subfamily G member 1 |
| chr1_+_214603173 | 0.79 |
ENST00000366955.8
|
CENPF
|
centromere protein F |
| chr11_-_66958366 | 0.77 |
ENST00000651036.1
ENST00000652125.1 ENST00000531614.6 ENST00000524491.6 ENST00000529047.6 ENST00000393960.7 ENST00000393958.7 ENST00000528403.6 ENST00000651854.1 |
PC
|
pyruvate carboxylase |
| chr6_-_4079100 | 0.76 |
ENST00000492651.5
ENST00000498677.5 ENST00000274673.8 |
FAM217A
|
family with sequence similarity 217 member A |
| chr18_+_12407896 | 0.75 |
ENST00000590956.5
ENST00000336990.8 ENST00000440960.6 ENST00000588729.5 |
PRELID3A
|
PRELI domain containing 3A |
| chr1_-_25906931 | 0.73 |
ENST00000357865.6
|
STMN1
|
stathmin 1 |
| chr21_-_42879516 | 0.71 |
ENST00000330317.6
ENST00000398208.3 |
WDR4
|
WD repeat domain 4 |
| chr16_-_4538819 | 0.71 |
ENST00000564828.5
|
CDIP1
|
cell death inducing p53 target 1 |
| chr6_+_151240368 | 0.68 |
ENST00000253332.5
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr4_-_102345196 | 0.62 |
ENST00000683412.1
ENST00000682227.1 |
SLC39A8
|
solute carrier family 39 member 8 |
| chr6_+_41921491 | 0.61 |
ENST00000230340.9
|
BYSL
|
bystin like |
| chr7_-_122699108 | 0.61 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr1_-_25905989 | 0.60 |
ENST00000399728.5
|
STMN1
|
stathmin 1 |
| chr6_+_151325665 | 0.57 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr12_-_84912705 | 0.57 |
ENST00000679933.1
ENST00000680260.1 ENST00000551010.2 ENST00000679453.1 ENST00000681281.1 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr10_-_52772763 | 0.56 |
ENST00000675947.1
ENST00000674931.1 |
MBL2
|
mannose binding lectin 2 |
| chr12_-_57752265 | 0.56 |
ENST00000547281.5
ENST00000257904.11 ENST00000546489.5 ENST00000552388.1 |
CDK4
|
cyclin dependent kinase 4 |
| chr12_-_57752345 | 0.54 |
ENST00000551800.5
ENST00000549606.5 ENST00000312990.10 |
CDK4
|
cyclin dependent kinase 4 |
| chr21_-_26843012 | 0.54 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr4_-_102345469 | 0.54 |
ENST00000356736.5
ENST00000682932.1 |
SLC39A8
|
solute carrier family 39 member 8 |
| chr14_+_92323154 | 0.53 |
ENST00000532405.6
ENST00000676001.1 ENST00000531433.5 |
SLC24A4
|
solute carrier family 24 member 4 |
| chr21_-_26843063 | 0.53 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr22_+_20117734 | 0.52 |
ENST00000416427.5
ENST00000421656.5 ENST00000423859.5 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr17_+_37375974 | 0.52 |
ENST00000615133.2
ENST00000611038.4 |
C17orf78
|
chromosome 17 open reading frame 78 |
| chr11_+_125904467 | 0.52 |
ENST00000263576.11
ENST00000530414.5 ENST00000530129.6 |
DDX25
|
DEAD-box helicase 25 |
| chr2_-_171434763 | 0.51 |
ENST00000442778.5
ENST00000453846.5 ENST00000612742.5 |
METTL8
|
methyltransferase like 8 |
| chr9_-_34637800 | 0.51 |
ENST00000680730.1
ENST00000477726.1 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr4_-_56435581 | 0.51 |
ENST00000264220.6
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr9_-_34637719 | 0.51 |
ENST00000378892.5
ENST00000680277.1 ENST00000277010.9 ENST00000679597.1 ENST00000680244.1 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr16_-_4538761 | 0.51 |
ENST00000567695.6
ENST00000562334.5 ENST00000562579.5 ENST00000563507.5 |
CDIP1
|
cell death inducing p53 target 1 |
| chr17_+_48892761 | 0.50 |
ENST00000355938.9
ENST00000393366.7 ENST00000503641.5 ENST00000514808.5 ENST00000506855.1 |
ATP5MC1
|
ATP synthase membrane subunit c locus 1 |
| chr1_-_109740304 | 0.50 |
ENST00000540225.2
|
GSTM3
|
glutathione S-transferase mu 3 |
| chr5_+_150357629 | 0.49 |
ENST00000650162.1
ENST00000377797.7 ENST00000445265.6 ENST00000323668.11 ENST00000643257.2 ENST00000646961.1 ENST00000513538.2 ENST00000439160.6 ENST00000394269.7 ENST00000427724.7 ENST00000504761.6 ENST00000513346.5 ENST00000515516.1 |
TCOF1
|
treacle ribosome biogenesis factor 1 |
| chr15_-_74203172 | 0.49 |
ENST00000616000.4
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr16_-_4538469 | 0.49 |
ENST00000588381.1
ENST00000563332.6 |
CDIP1
|
cell death inducing p53 target 1 |
| chr12_-_103841210 | 0.49 |
ENST00000392876.8
|
NT5DC3
|
5'-nucleotidase domain containing 3 |
| chr12_-_24949026 | 0.48 |
ENST00000539780.5
ENST00000546285.1 ENST00000342945.9 ENST00000261192.12 |
BCAT1
|
branched chain amino acid transaminase 1 |
| chr4_+_41256921 | 0.48 |
ENST00000284440.9
ENST00000508768.5 ENST00000512788.1 |
UCHL1
|
ubiquitin C-terminal hydrolase L1 |
| chrX_+_51332805 | 0.48 |
ENST00000356450.3
|
NUDT10
|
nudix hydrolase 10 |
| chr22_+_24594781 | 0.47 |
ENST00000456869.5
ENST00000411974.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_+_112645930 | 0.47 |
ENST00000272542.8
|
SLC20A1
|
solute carrier family 20 member 1 |
| chr2_-_50347710 | 0.46 |
ENST00000342183.9
ENST00000401710.5 |
NRXN1
|
neurexin 1 |
| chr9_-_128128102 | 0.46 |
ENST00000617202.4
|
PTGES2
|
prostaglandin E synthase 2 |
| chr4_-_53365976 | 0.46 |
ENST00000401642.8
ENST00000388940.8 ENST00000503450.1 |
SCFD2
|
sec1 family domain containing 2 |
| chr2_+_131011683 | 0.45 |
ENST00000355771.7
|
ARHGEF4
|
Rho guanine nucleotide exchange factor 4 |
| chr1_-_15585015 | 0.45 |
ENST00000375826.4
|
AGMAT
|
agmatinase |
| chr19_+_46303599 | 0.45 |
ENST00000300862.7
|
HIF3A
|
hypoxia inducible factor 3 subunit alpha |
| chrX_-_51496572 | 0.44 |
ENST00000375992.4
|
NUDT11
|
nudix hydrolase 11 |
| chr3_+_99817849 | 0.44 |
ENST00000421999.8
|
CMSS1
|
cms1 ribosomal small subunit homolog |
| chrX_-_101348676 | 0.44 |
ENST00000372902.4
ENST00000644112.2 |
TIMM8A
|
translocase of inner mitochondrial membrane 8A |
| chr5_+_111092329 | 0.43 |
ENST00000513710.4
|
WDR36
|
WD repeat domain 36 |
| chr1_+_116754422 | 0.43 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr16_+_8720706 | 0.42 |
ENST00000425191.6
ENST00000569156.5 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr2_-_219253909 | 0.42 |
ENST00000248437.9
|
TUBA4A
|
tubulin alpha 4a |
| chr5_+_111092172 | 0.41 |
ENST00000612402.4
|
WDR36
|
WD repeat domain 36 |
| chr8_+_38030496 | 0.41 |
ENST00000338825.5
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr11_-_124445696 | 0.41 |
ENST00000642064.1
|
OR8B8
|
olfactory receptor family 8 subfamily B member 8 |
| chr1_+_149782671 | 0.41 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chr4_+_127730386 | 0.40 |
ENST00000281154.6
|
SLC25A31
|
solute carrier family 25 member 31 |
| chr1_+_183023409 | 0.40 |
ENST00000258341.5
|
LAMC1
|
laminin subunit gamma 1 |
| chr17_-_4555371 | 0.40 |
ENST00000254718.9
ENST00000381556.6 |
MYBBP1A
|
MYB binding protein 1a |
| chr11_+_73218274 | 0.40 |
ENST00000393597.7
ENST00000311131.6 |
P2RY2
|
purinergic receptor P2Y2 |
| chr15_-_74202742 | 0.40 |
ENST00000395105.9
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr11_+_73218357 | 0.40 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y2 |
| chr1_-_111449209 | 0.39 |
ENST00000235090.10
|
WDR77
|
WD repeat domain 77 |
| chrX_-_54798253 | 0.39 |
ENST00000218436.7
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family member 6 |
| chr2_+_215312028 | 0.39 |
ENST00000236959.14
ENST00000435675.5 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr3_+_119147375 | 0.39 |
ENST00000490594.2
|
ENSG00000251012.2
|
novel chromosome 3 open reading frame 30 (C3orf30) and uroplakin 1B (UPK1B) |
| chr5_-_151157722 | 0.39 |
ENST00000517486.5
ENST00000377751.9 ENST00000521512.5 ENST00000517757.5 ENST00000354546.10 |
ANXA6
|
annexin A6 |
| chr11_-_64246907 | 0.38 |
ENST00000309318.8
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr17_+_17042433 | 0.38 |
ENST00000651222.2
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr17_-_75289443 | 0.38 |
ENST00000375261.8
ENST00000580273.1 |
SLC25A19
|
solute carrier family 25 member 19 |
| chr21_-_43659460 | 0.38 |
ENST00000443485.1
ENST00000291560.7 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr16_-_88785210 | 0.37 |
ENST00000301015.14
|
PIEZO1
|
piezo type mechanosensitive ion channel component 1 |
| chr19_+_45499797 | 0.37 |
ENST00000401593.5
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr16_+_56669832 | 0.37 |
ENST00000569155.1
|
MT1H
|
metallothionein 1H |
| chr10_-_69409275 | 0.37 |
ENST00000373307.5
|
TACR2
|
tachykinin receptor 2 |
| chr8_+_6708626 | 0.37 |
ENST00000285518.11
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr6_-_43528867 | 0.36 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr3_+_123067016 | 0.36 |
ENST00000316218.12
|
PDIA5
|
protein disulfide isomerase family A member 5 |
| chrX_+_15500800 | 0.36 |
ENST00000348343.11
|
BMX
|
BMX non-receptor tyrosine kinase |
| chrX_+_24693879 | 0.36 |
ENST00000379068.8
ENST00000677890.1 ENST00000379059.7 |
POLA1
|
DNA polymerase alpha 1, catalytic subunit |
| chr22_+_21642287 | 0.36 |
ENST00000248958.5
|
SDF2L1
|
stromal cell derived factor 2 like 1 |
| chr15_-_101295209 | 0.36 |
ENST00000254193.11
ENST00000626000.1 |
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr6_-_30744537 | 0.35 |
ENST00000259874.6
ENST00000376377.2 |
IER3
|
immediate early response 3 |
| chr1_+_38012706 | 0.35 |
ENST00000373014.5
|
UTP11
|
UTP11 small subunit processome component |
| chr4_+_56436131 | 0.35 |
ENST00000399688.7
|
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr5_+_136058849 | 0.35 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr22_-_41947087 | 0.34 |
ENST00000407253.7
ENST00000215980.10 |
CENPM
|
centromere protein M |
| chr17_+_80545422 | 0.34 |
ENST00000544334.6
|
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr13_+_50909905 | 0.34 |
ENST00000644034.1
ENST00000645955.1 |
RNASEH2B
|
ribonuclease H2 subunit B |
| chr6_-_31795627 | 0.34 |
ENST00000375663.8
|
VARS1
|
valyl-tRNA synthetase 1 |
| chr19_+_45499610 | 0.33 |
ENST00000396735.6
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr10_+_68956158 | 0.33 |
ENST00000354185.9
|
DDX21
|
DExD-box helicase 21 |
| chr2_+_17539964 | 0.33 |
ENST00000457525.5
|
VSNL1
|
visinin like 1 |
| chr14_+_64388296 | 0.33 |
ENST00000554739.5
ENST00000554768.6 ENST00000652179.1 ENST00000652337.1 ENST00000557370.3 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 |
| chr4_-_826092 | 0.33 |
ENST00000505203.1
|
CPLX1
|
complexin 1 |
| chr19_-_48511793 | 0.33 |
ENST00000600059.6
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr1_-_53142617 | 0.33 |
ENST00000371491.4
ENST00000371494.9 |
SLC1A7
|
solute carrier family 1 member 7 |
| chr3_-_50292404 | 0.32 |
ENST00000417626.8
|
IFRD2
|
interferon related developmental regulator 2 |
| chr21_-_30497160 | 0.32 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr17_+_42833390 | 0.32 |
ENST00000590720.6
ENST00000586114.5 ENST00000585805.5 ENST00000441946.6 ENST00000591152.5 ENST00000589469.5 ENST00000293362.7 ENST00000592169.2 |
PSME3
|
proteasome activator subunit 3 |
| chr18_-_35497591 | 0.32 |
ENST00000589273.1
ENST00000586489.5 |
INO80C
|
INO80 complex subunit C |
| chr17_-_50397472 | 0.32 |
ENST00000576448.1
ENST00000225972.8 |
LRRC59
|
leucine rich repeat containing 59 |
| chr12_+_64404338 | 0.32 |
ENST00000332707.10
|
XPOT
|
exportin for tRNA |
| chr19_-_49325181 | 0.31 |
ENST00000454748.7
ENST00000335875.9 ENST00000598828.1 |
SLC6A16
|
solute carrier family 6 member 16 |
| chr19_+_45498439 | 0.31 |
ENST00000451287.7
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr9_+_127397129 | 0.31 |
ENST00000610552.4
|
SLC2A8
|
solute carrier family 2 member 8 |
| chr2_+_157257687 | 0.31 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr9_+_127397153 | 0.31 |
ENST00000451404.5
ENST00000373371.8 |
SLC2A8
|
solute carrier family 2 member 8 |
| chr4_+_56435730 | 0.31 |
ENST00000514888.5
ENST00000264221.6 ENST00000505164.5 |
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr1_-_173824856 | 0.30 |
ENST00000682279.1
|
CENPL
|
centromere protein L |
| chr5_+_156326735 | 0.30 |
ENST00000435422.7
|
SGCD
|
sarcoglycan delta |
| chr9_+_109780292 | 0.30 |
ENST00000374530.7
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr22_+_39994926 | 0.30 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F |
| chr1_-_231241090 | 0.30 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr13_-_20192928 | 0.30 |
ENST00000382848.5
|
GJB2
|
gap junction protein beta 2 |
| chr1_-_52366124 | 0.30 |
ENST00000371586.6
ENST00000284376.8 |
CC2D1B
|
coiled-coil and C2 domain containing 1B |
| chr1_+_173824626 | 0.30 |
ENST00000648960.1
ENST00000648807.1 ENST00000649067.1 ENST00000649689.2 |
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr2_-_105396943 | 0.30 |
ENST00000409807.5
|
FHL2
|
four and a half LIM domains 2 |
| chr2_-_224402097 | 0.30 |
ENST00000409685.4
|
FAM124B
|
family with sequence similarity 124 member B |
| chr10_-_49539015 | 0.29 |
ENST00000681659.1
ENST00000680107.1 |
ERCC6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr5_+_161848112 | 0.29 |
ENST00000393943.10
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr19_+_17933001 | 0.29 |
ENST00000445755.7
|
CCDC124
|
coiled-coil domain containing 124 |
| chr21_+_36699100 | 0.29 |
ENST00000290399.11
|
SIM2
|
SIM bHLH transcription factor 2 |
| chr12_-_121802886 | 0.29 |
ENST00000545885.5
ENST00000542933.5 ENST00000428029.6 ENST00000541694.5 ENST00000536662.5 ENST00000535643.5 ENST00000541657.5 |
LINC01089
RHOF
|
long intergenic non-protein coding RNA 1089 ras homolog family member F, filopodia associated |
| chr10_-_49539112 | 0.29 |
ENST00000355832.10
ENST00000447839.7 |
ERCC6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr8_+_22995831 | 0.29 |
ENST00000522948.5
|
RHOBTB2
|
Rho related BTB domain containing 2 |
| chr18_-_31760864 | 0.29 |
ENST00000269205.7
ENST00000672005.1 |
SLC25A52
|
solute carrier family 25 member 52 |
| chr13_+_50909983 | 0.29 |
ENST00000643682.1
ENST00000642721.1 ENST00000616907.2 |
RNASEH2B
|
ribonuclease H2 subunit B |
| chr8_+_31033782 | 0.28 |
ENST00000298139.7
|
WRN
|
WRN RecQ like helicase |
| chr11_+_76783349 | 0.28 |
ENST00000333090.5
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr16_+_56669807 | 0.28 |
ENST00000332374.5
|
MT1H
|
metallothionein 1H |
| chr1_-_212699817 | 0.28 |
ENST00000243440.2
|
BATF3
|
basic leucine zipper ATF-like transcription factor 3 |
| chr2_+_172556039 | 0.28 |
ENST00000410055.5
ENST00000282077.8 |
PDK1
|
pyruvate dehydrogenase kinase 1 |
| chr15_-_82647503 | 0.28 |
ENST00000567678.1
ENST00000620182.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr11_+_57181945 | 0.28 |
ENST00000497933.3
|
LRRC55
|
leucine rich repeat containing 55 |
| chr21_-_5154527 | 0.27 |
ENST00000634020.1
ENST00000617716.4 |
ENSG00000275464.5
|
novel protein, similar to PWP2 periodic tryptophan protein homolog (yeast) PWP2 |
| chr14_+_22226711 | 0.27 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr4_-_102345061 | 0.27 |
ENST00000394833.6
|
SLC39A8
|
solute carrier family 39 member 8 |
| chr3_-_53844617 | 0.27 |
ENST00000481668.5
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr8_+_10672623 | 0.27 |
ENST00000304519.10
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr7_+_2244476 | 0.27 |
ENST00000397049.2
|
NUDT1
|
nudix hydrolase 1 |
| chr6_-_34146080 | 0.27 |
ENST00000538487.7
ENST00000374181.8 |
GRM4
|
glutamate metabotropic receptor 4 |
| chr2_-_197499857 | 0.27 |
ENST00000428204.6
ENST00000678170.1 ENST00000676933.1 ENST00000678621.1 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr16_+_6483379 | 0.27 |
ENST00000552089.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr9_+_127397184 | 0.27 |
ENST00000419917.5
ENST00000373352.5 ENST00000373360.7 |
SLC2A8
|
solute carrier family 2 member 8 |
| chr14_+_32934383 | 0.27 |
ENST00000551634.6
|
NPAS3
|
neuronal PAS domain protein 3 |
| chr1_-_205775182 | 0.27 |
ENST00000446390.6
|
RAB29
|
RAB29, member RAS oncogene family |
| chr1_-_53142577 | 0.26 |
ENST00000620347.5
ENST00000611397.5 |
SLC1A7
|
solute carrier family 1 member 7 |
| chr2_-_224401994 | 0.26 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124 member B |
| chr14_+_64704380 | 0.26 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr10_+_68956436 | 0.26 |
ENST00000620315.1
|
DDX21
|
DExD-box helicase 21 |
| chr1_+_173824694 | 0.26 |
ENST00000647645.1
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr6_+_44223770 | 0.26 |
ENST00000652453.1
ENST00000393841.6 ENST00000371724.6 ENST00000642777.1 ENST00000645692.1 |
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr11_+_62190212 | 0.25 |
ENST00000306238.3
|
SCGB1D1
|
secretoglobin family 1D member 1 |
| chr17_-_5468951 | 0.25 |
ENST00000225296.8
|
DHX33
|
DEAH-box helicase 33 |
| chr6_+_44219595 | 0.25 |
ENST00000393844.7
ENST00000652680.1 ENST00000643028.2 ENST00000371713.6 |
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr2_-_29074515 | 0.25 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr17_-_82098223 | 0.25 |
ENST00000306749.4
ENST00000635197.1 |
FASN
|
fatty acid synthase |
| chr6_-_88963409 | 0.25 |
ENST00000369475.7
ENST00000538899.2 |
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr20_+_2652622 | 0.25 |
ENST00000329276.10
ENST00000445139.1 |
NOP56
|
NOP56 ribonucleoprotein |
| chr6_+_44219527 | 0.25 |
ENST00000651428.1
|
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr8_-_70071226 | 0.25 |
ENST00000276594.3
|
PRDM14
|
PR/SET domain 14 |
| chr5_+_156327156 | 0.24 |
ENST00000337851.9
|
SGCD
|
sarcoglycan delta |
| chr16_-_57802401 | 0.24 |
ENST00000569112.5
ENST00000445690.7 ENST00000562311.5 ENST00000379655.8 |
KIFC3
|
kinesin family member C3 |
| chr7_-_44123508 | 0.24 |
ENST00000406581.6
ENST00000452185.5 ENST00000436844.5 ENST00000610533.6 ENST00000418438.1 |
POLD2
|
DNA polymerase delta 2, accessory subunit |
| chr16_+_83968244 | 0.24 |
ENST00000305202.9
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr17_-_75289212 | 0.24 |
ENST00000582778.1
ENST00000581988.5 ENST00000579207.5 ENST00000583332.5 ENST00000442286.6 ENST00000580151.5 ENST00000580994.5 ENST00000584438.1 ENST00000416858.7 ENST00000320362.7 |
SLC25A19
|
solute carrier family 25 member 19 |
| chrX_+_107628428 | 0.24 |
ENST00000643795.2
ENST00000372418.4 ENST00000646815.1 ENST00000372435.10 ENST00000372419.3 ENST00000676092.1 |
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr8_+_11802667 | 0.24 |
ENST00000443614.6
ENST00000220584.9 ENST00000525900.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr3_+_187197486 | 0.24 |
ENST00000312295.5
|
RTP1
|
receptor transporter protein 1 |
| chr1_+_20290869 | 0.24 |
ENST00000289815.13
ENST00000375079.6 |
VWA5B1
|
von Willebrand factor A domain containing 5B1 |
| chr1_+_154983318 | 0.23 |
ENST00000292180.8
ENST00000368433.5 ENST00000315144.14 ENST00000368432.5 ENST00000368431.7 |
FLAD1
|
flavin adenine dinucleotide synthetase 1 |
| chr17_+_7306975 | 0.23 |
ENST00000336452.11
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr21_+_44107380 | 0.23 |
ENST00000291576.12
|
PWP2
|
PWP2 small subunit processome component |
| chr19_+_1275997 | 0.23 |
ENST00000469144.5
|
FAM174C
|
family with sequence similarity 174 member C |
| chr5_+_110738983 | 0.23 |
ENST00000355943.8
ENST00000447245.6 |
SLC25A46
|
solute carrier family 25 member 46 |
| chr1_-_151993822 | 0.23 |
ENST00000368811.8
|
S100A10
|
S100 calcium binding protein A10 |
| chr12_+_57229694 | 0.23 |
ENST00000557487.5
ENST00000328923.8 ENST00000555634.5 ENST00000556689.5 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr4_+_74158099 | 0.23 |
ENST00000395759.6
ENST00000325278.7 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2 like |
| chr1_+_155610218 | 0.23 |
ENST00000649846.1
ENST00000245564.8 ENST00000368341.8 |
MSTO1
|
misato mitochondrial distribution and morphology regulator 1 |
| chr11_-_32435529 | 0.23 |
ENST00000448076.9
ENST00000452863.10 |
WT1
|
WT1 transcription factor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | GO:1905098 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.4 | 1.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 1.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.3 | 3.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.3 | 1.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.2 | 0.6 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.2 | 1.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 0.8 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 1.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 0.9 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.2 | 1.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.5 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.9 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.9 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.4 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 1.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 1.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.4 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.4 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.4 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.4 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.2 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.3 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 0.2 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.1 | 0.2 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.1 | 0.4 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.3 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 1.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.1 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.2 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 0.3 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 1.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.2 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.4 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.4 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 1.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.3 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.0 | 0.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.6 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 2.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.5 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.3 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 1.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.0 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.0 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.5 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.0 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.1 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.5 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.0 | GO:1901389 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.0 | 0.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 1.7 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 1.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 1.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 0.8 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.4 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 0.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.5 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.1 | 0.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 3.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:0046696 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.3 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 2.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0036379 | myofilament(GO:0036379) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.0 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.2 | 0.7 | GO:0004639 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 0.6 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.2 | 0.9 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.4 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.8 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.4 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.1 | 0.4 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.4 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.5 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.6 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.4 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0036219 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.2 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.1 | 0.5 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 1.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.8 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 3.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 2.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.7 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 2.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.6 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 4.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |