Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
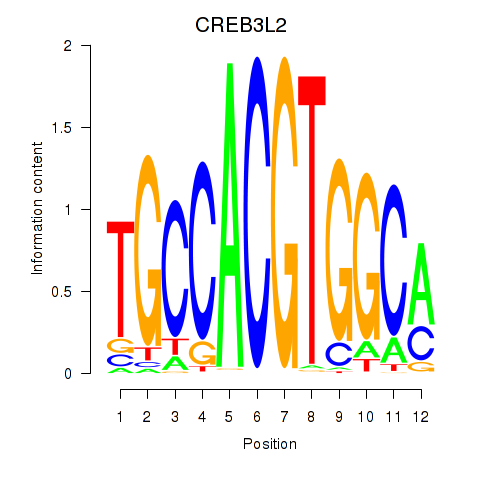
Results for CREB3L2
Z-value: 0.89
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.15 | CREB3L2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L2 | hg38_v1_chr7_-_138002017_138002138 | -0.46 | 9.6e-03 | Click! |
Activity profile of CREB3L2 motif
Sorted Z-values of CREB3L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_151240368 | 4.26 |
ENST00000253332.5
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr1_-_112956063 | 3.87 |
ENST00000538576.5
ENST00000369626.8 ENST00000458229.6 |
SLC16A1
|
solute carrier family 16 member 1 |
| chr21_-_26843012 | 3.65 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 3.59 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr6_+_151239951 | 2.97 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr17_+_41812974 | 2.19 |
ENST00000321562.9
|
FKBP10
|
FKBP prolyl isomerase 10 |
| chr20_+_25248036 | 2.11 |
ENST00000216962.9
|
PYGB
|
glycogen phosphorylase B |
| chr21_-_6467509 | 2.06 |
ENST00000624406.3
ENST00000398168.5 ENST00000624934.3 |
CBSL
|
cystathionine beta-synthase like |
| chr8_+_38030496 | 2.05 |
ENST00000338825.5
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr21_-_43075831 | 1.98 |
ENST00000398158.5
ENST00000398165.8 |
CBS
|
cystathionine beta-synthase |
| chr21_-_43076362 | 1.97 |
ENST00000359624.7
ENST00000352178.9 |
CBS
|
cystathionine beta-synthase |
| chr21_-_6468040 | 1.90 |
ENST00000618024.4
ENST00000617706.4 |
CBSL
|
cystathionine beta-synthase like |
| chr6_-_144008364 | 1.58 |
ENST00000625622.2
|
PLAGL1
|
PLAG1 like zinc finger 1 |
| chr1_-_11060000 | 1.51 |
ENST00000376957.7
|
SRM
|
spermidine synthase |
| chr19_-_2783241 | 1.40 |
ENST00000676943.1
ENST00000589251.5 ENST00000221566.7 ENST00000676984.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr19_-_2783308 | 1.39 |
ENST00000677562.1
ENST00000677754.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chrX_+_55220385 | 1.38 |
ENST00000374952.1
|
PAGE5
|
PAGE family member 5 |
| chr6_-_144008118 | 1.37 |
ENST00000629195.2
ENST00000650125.1 ENST00000647880.1 ENST00000649307.1 ENST00000674357.1 ENST00000417959.4 ENST00000635591.1 |
PLAGL1
HYMAI
|
PLAG1 like zinc finger 1 hydatidiform mole associated and imprinted |
| chr3_+_123067016 | 1.36 |
ENST00000316218.12
|
PDIA5
|
protein disulfide isomerase family A member 5 |
| chr1_-_68497030 | 1.32 |
ENST00000456315.7
ENST00000525124.1 ENST00000370966.9 |
DEPDC1
|
DEP domain containing 1 |
| chr6_-_144008396 | 1.29 |
ENST00000354765.6
ENST00000416623.5 ENST00000649211.1 |
PLAGL1
|
PLAG1 like zinc finger 1 |
| chr12_+_57229694 | 1.25 |
ENST00000557487.5
ENST00000328923.8 ENST00000555634.5 ENST00000556689.5 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr2_-_46916020 | 1.20 |
ENST00000409800.5
ENST00000409218.5 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr7_+_2631978 | 1.19 |
ENST00000258796.12
|
TTYH3
|
tweety family member 3 |
| chr2_-_46915745 | 1.10 |
ENST00000649435.1
ENST00000409105.5 ENST00000319466.9 ENST00000409973.5 ENST00000409913.5 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr16_+_2520339 | 1.10 |
ENST00000568263.5
ENST00000302956.8 ENST00000293971.11 ENST00000413459.7 ENST00000648227.1 ENST00000566706.5 ENST00000569879.5 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr2_+_64453969 | 1.09 |
ENST00000464281.5
|
LGALSL
|
galectin like |
| chr17_-_38853629 | 1.00 |
ENST00000378096.3
ENST00000479035.7 ENST00000394332.5 ENST00000394333.5 ENST00000577407.5 |
RPL23
|
ribosomal protein L23 |
| chr12_+_57230301 | 0.99 |
ENST00000553474.5
|
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr2_+_200811882 | 0.95 |
ENST00000409600.6
|
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr12_+_57230086 | 0.93 |
ENST00000414700.7
ENST00000557703.5 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr9_+_706841 | 0.91 |
ENST00000382293.7
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chrX_+_154458274 | 0.90 |
ENST00000369682.4
|
PLXNA3
|
plexin A3 |
| chr2_-_69387130 | 0.90 |
ENST00000674438.1
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr20_-_63537361 | 0.90 |
ENST00000217185.3
ENST00000542869.3 |
PTK6
|
protein tyrosine kinase 6 |
| chr12_+_57230336 | 0.90 |
ENST00000555773.5
ENST00000554975.5 ENST00000449049.7 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr17_-_76726590 | 0.86 |
ENST00000397625.9
ENST00000445478.6 |
JMJD6
|
jumonji domain containing 6, arginine demethylase and lysine hydroxylase |
| chr8_+_22367526 | 0.85 |
ENST00000289952.9
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 member 14 |
| chr2_+_64454145 | 0.84 |
ENST00000238875.10
|
LGALSL
|
galectin like |
| chr2_+_112645930 | 0.84 |
ENST00000272542.8
|
SLC20A1
|
solute carrier family 20 member 1 |
| chr12_-_47758828 | 0.82 |
ENST00000389212.7
ENST00000449771.7 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr17_-_76726753 | 0.82 |
ENST00000617192.4
|
JMJD6
|
jumonji domain containing 6, arginine demethylase and lysine hydroxylase |
| chr17_-_41812586 | 0.82 |
ENST00000355468.7
ENST00000590496.1 |
P3H4
|
prolyl 3-hydroxylase family member 4 (inactive) |
| chr7_-_1556194 | 0.81 |
ENST00000297477.10
|
TMEM184A
|
transmembrane protein 184A |
| chr22_-_50085414 | 0.80 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr15_+_88638947 | 0.80 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr17_+_77405070 | 0.79 |
ENST00000585930.5
|
SEPTIN9
|
septin 9 |
| chr22_-_50085331 | 0.77 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr3_-_123884290 | 0.75 |
ENST00000346322.9
ENST00000360772.7 ENST00000360304.8 |
MYLK
|
myosin light chain kinase |
| chr1_+_52602347 | 0.75 |
ENST00000361314.5
|
GPX7
|
glutathione peroxidase 7 |
| chr2_-_27119012 | 0.75 |
ENST00000312734.8
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr17_+_49132763 | 0.74 |
ENST00000393354.7
|
B4GALNT2
|
beta-1,4-N-acetyl-galactosaminyltransferase 2 |
| chr1_-_151008365 | 0.74 |
ENST00000361936.9
ENST00000361738.11 |
MINDY1
|
MINDY lysine 48 deubiquitinase 1 |
| chr20_+_62952674 | 0.74 |
ENST00000370349.7
ENST00000370351.9 |
SLC17A9
|
solute carrier family 17 member 9 |
| chr19_+_48954850 | 0.73 |
ENST00000345358.12
ENST00000539787.2 ENST00000415969.6 ENST00000354470.7 ENST00000506183.5 ENST00000391871.4 ENST00000293288.12 |
BAX
|
BCL2 associated X, apoptosis regulator |
| chr8_+_22367259 | 0.72 |
ENST00000520644.1
ENST00000359741.10 ENST00000381237.6 ENST00000240095.10 |
SLC39A14
|
solute carrier family 39 member 14 |
| chr2_-_27119099 | 0.72 |
ENST00000402550.5
ENST00000402394.6 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr3_+_141426108 | 0.71 |
ENST00000441582.2
ENST00000510726.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr15_+_62066975 | 0.70 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium dependent domain containing 4A |
| chr2_+_79512993 | 0.68 |
ENST00000496558.5
ENST00000451966.5 ENST00000402739.9 ENST00000629316.2 |
CTNNA2
|
catenin alpha 2 |
| chr1_-_109619605 | 0.68 |
ENST00000679935.1
|
GNAT2
|
G protein subunit alpha transducin 2 |
| chr12_+_120978537 | 0.65 |
ENST00000257555.11
ENST00000400024.6 |
HNF1A
|
HNF1 homeobox A |
| chr15_+_88639009 | 0.64 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr1_-_11805924 | 0.64 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr3_-_57597443 | 0.64 |
ENST00000463880.1
|
ARF4
|
ADP ribosylation factor 4 |
| chr1_+_236686454 | 0.64 |
ENST00000542672.6
ENST00000366578.6 ENST00000682015.1 ENST00000651275.1 |
ACTN2
|
actinin alpha 2 |
| chr17_-_18363451 | 0.64 |
ENST00000354098.7
|
SHMT1
|
serine hydroxymethyltransferase 1 |
| chr12_+_120978686 | 0.64 |
ENST00000541395.5
ENST00000544413.2 |
HNF1A
|
HNF1 homeobox A |
| chr5_-_55712280 | 0.63 |
ENST00000506624.5
ENST00000513275.5 ENST00000513993.5 ENST00000396865.7 ENST00000503891.5 ENST00000507109.5 |
SLC38A9
|
solute carrier family 38 member 9 |
| chr2_+_200812185 | 0.63 |
ENST00000409226.5
ENST00000452790.6 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr17_-_76726453 | 0.62 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6, arginine demethylase and lysine hydroxylase |
| chr19_-_9792991 | 0.61 |
ENST00000592587.1
|
ZNF846
|
zinc finger protein 846 |
| chrX_-_120560884 | 0.60 |
ENST00000404115.8
|
CUL4B
|
cullin 4B |
| chr1_-_33431079 | 0.60 |
ENST00000683057.1
|
PHC2
|
polyhomeotic homolog 2 |
| chr2_+_176092715 | 0.60 |
ENST00000392539.4
|
HOXD13
|
homeobox D13 |
| chrX_+_55220338 | 0.59 |
ENST00000374955.8
ENST00000289619.9 |
PAGE5
|
PAGE family member 5 |
| chr22_+_44172932 | 0.59 |
ENST00000422871.5
|
PARVG
|
parvin gamma |
| chrX_+_48574938 | 0.58 |
ENST00000376755.1
|
RBM3
|
RNA binding motif protein 3 |
| chr1_-_11805977 | 0.58 |
ENST00000376486.3
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr17_-_7234262 | 0.58 |
ENST00000575756.5
ENST00000575458.5 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr10_-_70170466 | 0.57 |
ENST00000373239.2
ENST00000373241.9 ENST00000373242.6 |
SAR1A
|
secretion associated Ras related GTPase 1A |
| chr19_-_4670331 | 0.56 |
ENST00000262947.8
ENST00000599630.1 |
MYDGF
|
myeloid derived growth factor |
| chrX_-_15854743 | 0.56 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr4_-_48906805 | 0.56 |
ENST00000620187.4
|
OCIAD2
|
OCIA domain containing 2 |
| chr7_-_134459089 | 0.56 |
ENST00000285930.9
|
AKR1B1
|
aldo-keto reductase family 1 member B |
| chrX_+_132023294 | 0.56 |
ENST00000481105.5
ENST00000354719.10 ENST00000394334.7 ENST00000394335.6 |
STK26
|
serine/threonine kinase 26 |
| chr6_+_159969070 | 0.55 |
ENST00000356956.6
|
IGF2R
|
insulin like growth factor 2 receptor |
| chr17_+_51166431 | 0.55 |
ENST00000393190.4
|
NME2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr17_-_42181116 | 0.54 |
ENST00000264661.4
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chr8_+_81280527 | 0.54 |
ENST00000297258.11
|
FABP5
|
fatty acid binding protein 5 |
| chr2_-_27118900 | 0.54 |
ENST00000405600.5
ENST00000260595.9 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr17_-_42181081 | 0.54 |
ENST00000607371.5
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chrY_-_21894439 | 0.53 |
ENST00000418956.2
ENST00000382680.5 |
RBMY1D
|
RNA binding motif protein Y-linked family 1 member D |
| chr14_+_49598761 | 0.53 |
ENST00000318317.8
|
LRR1
|
leucine rich repeat protein 1 |
| chr13_+_32315468 | 0.53 |
ENST00000530893.6
ENST00000380152.8 |
BRCA2
|
BRCA2 DNA repair associated |
| chr13_+_32315071 | 0.52 |
ENST00000544455.6
|
BRCA2
|
BRCA2 DNA repair associated |
| chr4_-_48906788 | 0.52 |
ENST00000273860.8
|
OCIAD2
|
OCIA domain containing 2 |
| chr14_+_104689588 | 0.52 |
ENST00000330634.11
ENST00000392634.9 ENST00000675482.1 ENST00000398337.8 |
INF2
|
inverted formin 2 |
| chr3_+_50617119 | 0.52 |
ENST00000430409.5
ENST00000621469.5 ENST00000357955.6 |
MAPKAPK3
|
MAPK activated protein kinase 3 |
| chrY_-_21918032 | 0.51 |
ENST00000382658.5
ENST00000382659.6 |
RBMY1E
|
RNA binding motif protein Y-linked family 1 member E |
| chrY_+_21534879 | 0.51 |
ENST00000382707.6
|
RBMY1A1
|
RNA binding motif protein Y-linked family 1 member A1 |
| chr15_+_58771280 | 0.51 |
ENST00000559228.6
ENST00000450403.3 |
MINDY2
|
MINDY lysine 48 deubiquitinase 2 |
| chr3_-_179071742 | 0.51 |
ENST00000311417.7
ENST00000652290.1 |
ZMAT3
|
zinc finger matrin-type 3 |
| chr4_+_187995764 | 0.51 |
ENST00000509524.5
ENST00000326866.5 |
ZFP42
|
ZFP42 zinc finger protein |
| chr5_-_151157722 | 0.50 |
ENST00000517486.5
ENST00000377751.9 ENST00000521512.5 ENST00000517757.5 ENST00000354546.10 |
ANXA6
|
annexin A6 |
| chrY_+_21511338 | 0.49 |
ENST00000383020.7
ENST00000619219.1 ENST00000439108.6 ENST00000303902.9 |
RBMY1B
RBMY1A1
|
RNA binding motif protein Y-linked family 1 member B RNA binding motif protein Y-linked family 1 member A1 |
| chr17_-_7234462 | 0.49 |
ENST00000005340.10
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr10_+_63133279 | 0.49 |
ENST00000277746.11
|
NRBF2
|
nuclear receptor binding factor 2 |
| chr9_-_131270493 | 0.48 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr10_+_117543567 | 0.48 |
ENST00000616794.1
|
EMX2
|
empty spiracles homeobox 2 |
| chr4_-_185425941 | 0.48 |
ENST00000264689.11
ENST00000505357.1 |
UFSP2
|
UFM1 specific peptidase 2 |
| chrX_-_149631682 | 0.48 |
ENST00000511776.1
ENST00000507237.5 |
TMEM185A
|
transmembrane protein 185A |
| chr3_+_189171948 | 0.48 |
ENST00000345063.8
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr5_-_159209503 | 0.48 |
ENST00000424310.7
ENST00000611185.4 |
RNF145
|
ring finger protein 145 |
| chr11_+_120210991 | 0.47 |
ENST00000328965.9
|
OAF
|
out at first homolog |
| chr4_-_48906720 | 0.46 |
ENST00000381464.6
ENST00000508632.6 |
OCIAD2
|
OCIA domain containing 2 |
| chr17_+_50373214 | 0.45 |
ENST00000393271.6
ENST00000338165.9 ENST00000511519.6 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr5_-_144170607 | 0.45 |
ENST00000448443.6
ENST00000513112.5 ENST00000519064.5 ENST00000274496.10 ENST00000522203.5 |
YIPF5
|
Yip1 domain family member 5 |
| chr8_-_98117155 | 0.45 |
ENST00000254878.8
ENST00000521560.1 |
RIDA
|
reactive intermediate imine deaminase A homolog |
| chr10_-_125775821 | 0.44 |
ENST00000368808.3
|
MMP21
|
matrix metallopeptidase 21 |
| chr2_-_15561305 | 0.44 |
ENST00000281513.10
|
NBAS
|
NBAS subunit of NRZ tethering complex |
| chr6_+_167111789 | 0.43 |
ENST00000400926.5
|
CCR6
|
C-C motif chemokine receptor 6 |
| chrX_-_120561424 | 0.42 |
ENST00000681206.1
ENST00000679927.1 ENST00000336592.11 |
CUL4B
|
cullin 4B |
| chrX_-_120560947 | 0.42 |
ENST00000674137.11
ENST00000371322.11 ENST00000681090.1 |
CUL4B
|
cullin 4B |
| chr8_-_98117110 | 0.42 |
ENST00000520507.5
|
RIDA
|
reactive intermediate imine deaminase A homolog |
| chr4_-_8127650 | 0.41 |
ENST00000545242.6
ENST00000676532.1 |
ABLIM2
|
actin binding LIM protein family member 2 |
| chr11_-_64246907 | 0.41 |
ENST00000309318.8
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr5_-_1799750 | 0.41 |
ENST00000510999.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr7_-_142885737 | 0.41 |
ENST00000359396.9
ENST00000436401.1 |
TRPV6
|
transient receptor potential cation channel subfamily V member 6 |
| chr18_+_9708230 | 0.40 |
ENST00000578921.6
|
RAB31
|
RAB31, member RAS oncogene family |
| chr10_-_70233420 | 0.39 |
ENST00000373232.8
ENST00000625364.1 |
PPA1
|
inorganic pyrophosphatase 1 |
| chr1_-_11805949 | 0.39 |
ENST00000376590.9
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr19_-_46787278 | 0.39 |
ENST00000412532.6
|
SLC1A5
|
solute carrier family 1 member 5 |
| chr17_-_65056659 | 0.39 |
ENST00000439174.7
|
GNA13
|
G protein subunit alpha 13 |
| chr9_-_127715602 | 0.39 |
ENST00000456267.5
ENST00000414832.2 |
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr10_+_102132994 | 0.38 |
ENST00000413464.6
ENST00000278070.7 |
PPRC1
|
PPARG related coactivator 1 |
| chr13_+_113297217 | 0.38 |
ENST00000332556.5
|
LAMP1
|
lysosomal associated membrane protein 1 |
| chr3_-_57079287 | 0.38 |
ENST00000338458.8
ENST00000468727.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr12_+_122980060 | 0.37 |
ENST00000543566.6
ENST00000453766.7 ENST00000392435.7 ENST00000413381.6 ENST00000426960.6 |
ARL6IP4
|
ADP ribosylation factor like GTPase 6 interacting protein 4 |
| chr7_-_6272639 | 0.37 |
ENST00000396741.3
|
CYTH3
|
cytohesin 3 |
| chr17_-_50373173 | 0.37 |
ENST00000225969.9
ENST00000503633.5 ENST00000442592.3 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr17_-_18363504 | 0.36 |
ENST00000583780.1
ENST00000316694.8 ENST00000352886.10 |
SHMT1
|
serine hydroxymethyltransferase 1 |
| chr4_+_127730386 | 0.36 |
ENST00000281154.6
|
SLC25A31
|
solute carrier family 25 member 31 |
| chr2_+_171687409 | 0.36 |
ENST00000452242.5
ENST00000340296.8 |
DYNC1I2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr20_-_45891200 | 0.36 |
ENST00000372518.5
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr14_+_93333210 | 0.36 |
ENST00000256339.8
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr1_-_77682639 | 0.35 |
ENST00000370801.8
ENST00000433749.5 |
ZZZ3
|
zinc finger ZZ-type containing 3 |
| chr18_+_158513 | 0.35 |
ENST00000400266.7
ENST00000580410.5 ENST00000261601.8 ENST00000383589.6 |
USP14
|
ubiquitin specific peptidase 14 |
| chr7_-_8262668 | 0.34 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1 |
| chr16_+_78202 | 0.34 |
ENST00000356432.8
ENST00000219431.4 |
MPG
|
N-methylpurine DNA glycosylase |
| chr8_+_1823918 | 0.34 |
ENST00000349830.8
|
ARHGEF10
|
Rho guanine nucleotide exchange factor 10 |
| chr5_+_177303768 | 0.34 |
ENST00000303204.9
ENST00000503216.5 |
PRELID1
|
PRELI domain containing 1 |
| chr2_-_73112885 | 0.33 |
ENST00000486777.7
|
RAB11FIP5
|
RAB11 family interacting protein 5 |
| chr21_+_43789522 | 0.33 |
ENST00000497547.2
|
RRP1
|
ribosomal RNA processing 1 |
| chr19_-_12919256 | 0.33 |
ENST00000293695.8
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chrX_-_149631787 | 0.33 |
ENST00000600449.8
ENST00000611119.4 |
TMEM185A
|
transmembrane protein 185A |
| chr16_+_70579867 | 0.32 |
ENST00000429149.6
|
IL34
|
interleukin 34 |
| chr2_+_206159580 | 0.32 |
ENST00000236957.9
ENST00000392221.5 ENST00000445505.5 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr16_+_24539536 | 0.32 |
ENST00000568015.5
ENST00000319715.10 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr5_-_1799818 | 0.31 |
ENST00000505059.7
ENST00000382647.7 |
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr7_-_6272575 | 0.31 |
ENST00000350796.8
|
CYTH3
|
cytohesin 3 |
| chr9_-_122093273 | 0.31 |
ENST00000321582.11
|
TTLL11
|
tubulin tyrosine ligase like 11 |
| chr2_+_206159884 | 0.30 |
ENST00000392222.7
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr5_+_41904329 | 0.30 |
ENST00000381647.7
ENST00000612065.1 |
C5orf51
|
chromosome 5 open reading frame 51 |
| chr5_-_139389905 | 0.29 |
ENST00000302125.9
|
MZB1
|
marginal zone B and B1 cell specific protein |
| chr2_+_215312028 | 0.29 |
ENST00000236959.14
ENST00000435675.5 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr14_-_105708627 | 0.29 |
ENST00000641837.1
ENST00000390547.3 |
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr8_+_1823967 | 0.29 |
ENST00000520359.5
ENST00000518288.5 |
ARHGEF10
|
Rho guanine nucleotide exchange factor 10 |
| chr1_-_1231966 | 0.29 |
ENST00000360001.12
|
SDF4
|
stromal cell derived factor 4 |
| chr12_-_121296685 | 0.28 |
ENST00000412367.6
ENST00000404169.8 ENST00000402834.8 |
CAMKK2
|
calcium/calmodulin dependent protein kinase kinase 2 |
| chr1_-_1232031 | 0.28 |
ENST00000263741.12
|
SDF4
|
stromal cell derived factor 4 |
| chr17_-_81937320 | 0.28 |
ENST00000577624.5
ENST00000403172.8 ENST00000619204.4 ENST00000629768.2 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr17_-_81937277 | 0.28 |
ENST00000405481.8
ENST00000329875.13 ENST00000585215.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr21_+_44455471 | 0.28 |
ENST00000291592.6
|
LRRC3
|
leucine rich repeat containing 3 |
| chr1_+_231241195 | 0.28 |
ENST00000436239.5
ENST00000366647.9 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr16_+_85027735 | 0.28 |
ENST00000258180.7
ENST00000538274.5 |
KIAA0513
|
KIAA0513 |
| chr17_+_77281429 | 0.27 |
ENST00000591198.5
ENST00000427177.6 |
SEPTIN9
|
septin 9 |
| chr17_-_81937221 | 0.27 |
ENST00000402252.6
ENST00000583564.5 ENST00000585244.1 ENST00000337943.9 ENST00000579698.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr2_+_171687501 | 0.26 |
ENST00000508530.5
|
DYNC1I2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr15_-_72375940 | 0.26 |
ENST00000567213.2
ENST00000566304.5 ENST00000567159.5 ENST00000683884.1 ENST00000684520.1 ENST00000268097.10 |
HEXA
|
hexosaminidase subunit alpha |
| chr20_+_62642492 | 0.26 |
ENST00000217159.6
|
SLCO4A1
|
solute carrier organic anion transporter family member 4A1 |
| chr22_+_38705922 | 0.26 |
ENST00000216044.10
|
GTPBP1
|
GTP binding protein 1 |
| chr1_+_63322558 | 0.26 |
ENST00000371116.4
|
FOXD3
|
forkhead box D3 |
| chr5_+_66144204 | 0.26 |
ENST00000612404.4
|
SREK1
|
splicing regulatory glutamic acid and lysine rich protein 1 |
| chr19_-_3500664 | 0.26 |
ENST00000427575.6
|
DOHH
|
deoxyhypusine hydroxylase |
| chr5_-_1799853 | 0.26 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr12_+_48328980 | 0.26 |
ENST00000335017.1
|
H1-7
|
H1.7 linker histone |
| chr5_+_92106 | 0.26 |
ENST00000637938.1
|
PLEKHG4B
|
pleckstrin homology and RhoGEF domain containing G4B |
| chrX_+_106693838 | 0.25 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr2_-_127885956 | 0.24 |
ENST00000272647.10
ENST00000680886.1 |
AMMECR1L
|
AMMECR1 like |
| chr5_+_144170843 | 0.24 |
ENST00000512467.6
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr5_+_178941186 | 0.24 |
ENST00000320129.7
ENST00000519564.2 |
ZNF454
|
zinc finger protein 454 |
| chr19_+_1275997 | 0.24 |
ENST00000469144.5
|
FAM174C
|
family with sequence similarity 174 member C |
| chr15_+_58431985 | 0.23 |
ENST00000433326.2
ENST00000299022.10 |
LIPC
|
lipase C, hepatic type |
| chr1_+_43979877 | 0.23 |
ENST00000356836.10
ENST00000309519.8 |
B4GALT2
|
beta-1,4-galactosyltransferase 2 |
| chr6_-_136550819 | 0.23 |
ENST00000616617.4
ENST00000618822.4 |
MAP7
|
microtubule associated protein 7 |
| chr14_+_95876385 | 0.23 |
ENST00000504119.1
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA |
| chr19_+_34172645 | 0.23 |
ENST00000586157.2
|
LSM14A
|
LSM14A mRNA processing body assembly factor |
| chr6_-_27872334 | 0.22 |
ENST00000616365.2
|
H3C11
|
H3 clustered histone 11 |
| chr1_-_1232061 | 0.22 |
ENST00000660930.1
|
SDF4
|
stromal cell derived factor 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.8 | 5.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.8 | 3.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.8 | 2.3 | GO:0070078 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine(GO:0018395) histone arginine demethylation(GO:0070077) histone H3-R2 demethylation(GO:0070078) histone H4-R3 demethylation(GO:0070079) |
| 0.6 | 7.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.5 | 3.9 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.4 | 1.3 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.4 | 2.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.4 | 7.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.4 | 1.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 1.6 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.3 | 1.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 0.8 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.2 | 1.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 0.7 | GO:1902512 | B cell selection(GO:0002339) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.2 | 0.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 1.5 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 0.6 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.2 | 1.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.2 | 0.6 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.2 | 0.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 1.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.5 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 1.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.4 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.4 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 1.6 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.1 | 0.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.6 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 1.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.3 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.4 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.8 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.1 | 0.9 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.7 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.3 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.3 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.9 | GO:1902074 | response to salt(GO:1902074) |
| 0.1 | 0.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 1.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 1.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.3 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 1.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 3.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 2.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 2.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.2 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 1.1 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.1 | 0.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0071750 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 0.1 | 0.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.2 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.8 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 6.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 2.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 6.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.5 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 2.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.8 | 5.1 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.8 | 2.3 | GO:0033749 | histone demethylase activity (H3-R2 specific)(GO:0033746) histone demethylase activity (H4-R3 specific)(GO:0033749) |
| 0.5 | 2.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 1.5 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.5 | 1.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 2.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.4 | 7.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 3.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 1.6 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 1.2 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 1.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 0.6 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.2 | 0.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.6 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.1 | 0.6 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.7 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.6 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 7.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.9 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.9 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 1.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 7.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 2.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |