Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
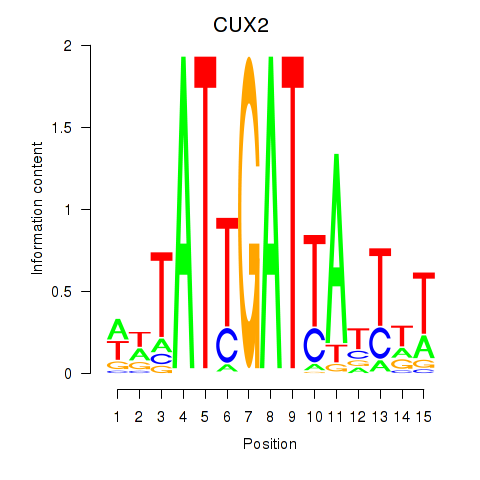
Results for CUX2
Z-value: 0.73
Transcription factors associated with CUX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX2
|
ENSG00000111249.14 | CUX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX2 | hg38_v1_chr12_+_111034136_111034173 | 0.08 | 6.7e-01 | Click! |
Activity profile of CUX2 motif
Sorted Z-values of CUX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_109075944 | 2.23 |
ENST00000338366.6
|
TAF13
|
TATA-box binding protein associated factor 13 |
| chr7_-_23470469 | 2.17 |
ENST00000258729.8
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr2_+_95025700 | 2.11 |
ENST00000309988.9
ENST00000353004.7 ENST00000354078.7 ENST00000349807.3 |
MAL
|
mal, T cell differentiation protein |
| chr1_+_31576485 | 1.82 |
ENST00000457433.6
ENST00000271064.12 |
TINAGL1
|
tubulointerstitial nephritis antigen like 1 |
| chr12_-_10826358 | 1.79 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr1_-_153041111 | 1.72 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr21_-_26843012 | 1.67 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr8_-_61646807 | 1.66 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr2_-_215393126 | 1.65 |
ENST00000456923.5
|
FN1
|
fibronectin 1 |
| chr21_-_26843063 | 1.58 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr15_-_59689283 | 1.56 |
ENST00000607373.6
ENST00000612191.4 ENST00000267859.8 |
BNIP2
|
BCL2 interacting protein 2 |
| chr8_-_90082871 | 1.52 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr9_+_102995308 | 1.45 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr3_+_100635598 | 1.42 |
ENST00000475887.1
|
ADGRG7
|
adhesion G protein-coupled receptor G7 |
| chr2_+_101839815 | 1.42 |
ENST00000421882.5
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr6_+_151239951 | 1.35 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr1_+_171185293 | 1.33 |
ENST00000209929.10
|
FMO2
|
flavin containing dimethylaniline monoxygenase 2 |
| chr2_-_191151568 | 1.29 |
ENST00000358470.8
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr7_+_80602200 | 1.27 |
ENST00000534394.5
|
CD36
|
CD36 molecule |
| chr7_+_80602150 | 1.24 |
ENST00000309881.11
|
CD36
|
CD36 molecule |
| chr16_+_11345429 | 1.17 |
ENST00000576027.1
ENST00000312499.6 ENST00000648619.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr8_+_53851786 | 1.16 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr11_-_105139750 | 1.14 |
ENST00000530950.2
|
CARD18
|
caspase recruitment domain family member 18 |
| chr9_+_122371036 | 1.10 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr19_+_47713412 | 1.07 |
ENST00000538399.1
ENST00000263277.8 |
EHD2
|
EH domain containing 2 |
| chr19_+_44905785 | 1.07 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr6_-_131000722 | 1.06 |
ENST00000528282.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chrX_-_108439472 | 1.05 |
ENST00000372216.8
|
COL4A6
|
collagen type IV alpha 6 chain |
| chr14_-_80959484 | 1.00 |
ENST00000555529.5
ENST00000556042.5 ENST00000556981.5 |
CEP128
|
centrosomal protein 128 |
| chr22_+_44752552 | 0.92 |
ENST00000389774.6
ENST00000356099.11 ENST00000396119.6 ENST00000336963.8 ENST00000412433.5 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr19_+_41797147 | 0.89 |
ENST00000596544.1
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr10_-_119536533 | 0.88 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr11_+_5389377 | 0.88 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr2_+_102337148 | 0.85 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr19_-_4535221 | 0.83 |
ENST00000381848.7
ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr11_+_62856149 | 0.82 |
ENST00000535296.5
|
SLC3A2
|
solute carrier family 3 member 2 |
| chr6_-_130956371 | 0.82 |
ENST00000639623.1
ENST00000525193.5 ENST00000527659.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr14_-_63728027 | 0.80 |
ENST00000247225.7
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr9_+_122371014 | 0.80 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr15_+_41286011 | 0.79 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr6_+_121435595 | 0.79 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr22_+_18527802 | 0.78 |
ENST00000612978.5
|
TMEM191B
|
transmembrane protein 191B |
| chr19_-_7058640 | 0.78 |
ENST00000333843.8
|
MBD3L3
|
methyl-CpG binding domain protein 3 like 3 |
| chr19_+_926001 | 0.76 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr17_-_41369807 | 0.73 |
ENST00000251646.8
|
KRT33B
|
keratin 33B |
| chr15_+_40382715 | 0.72 |
ENST00000416151.6
ENST00000249776.12 |
KNSTRN
|
kinetochore localized astrin (SPAG5) binding protein |
| chr19_+_41376560 | 0.72 |
ENST00000436170.6
|
TMEM91
|
transmembrane protein 91 |
| chr3_+_123067016 | 0.70 |
ENST00000316218.12
|
PDIA5
|
protein disulfide isomerase family A member 5 |
| chr5_+_163460650 | 0.69 |
ENST00000358715.3
|
HMMR
|
hyaluronan mediated motility receptor |
| chr19_+_41376499 | 0.69 |
ENST00000392002.7
|
TMEM91
|
transmembrane protein 91 |
| chr15_+_40382764 | 0.69 |
ENST00000448395.6
|
KNSTRN
|
kinetochore localized astrin (SPAG5) binding protein |
| chr19_+_45499797 | 0.68 |
ENST00000401593.5
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr6_+_26272923 | 0.66 |
ENST00000377733.4
|
H2BC10
|
H2B clustered histone 10 |
| chr5_+_163460623 | 0.66 |
ENST00000393915.9
ENST00000432118.6 |
HMMR
|
hyaluronan mediated motility receptor |
| chr6_-_27146841 | 0.66 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr5_-_11588842 | 0.65 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chr7_+_80646305 | 0.65 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr16_-_88663065 | 0.64 |
ENST00000301012.8
ENST00000569177.5 |
MVD
|
mevalonate diphosphate decarboxylase |
| chr6_-_49636832 | 0.64 |
ENST00000371175.10
ENST00000646272.1 ENST00000646939.1 ENST00000618248.3 ENST00000229810.9 ENST00000646963.1 |
RHAG
|
Rh associated glycoprotein |
| chr5_-_11589019 | 0.64 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr3_-_48089203 | 0.64 |
ENST00000468075.2
ENST00000360240.10 |
MAP4
|
microtubule associated protein 4 |
| chrX_+_54920796 | 0.64 |
ENST00000442098.5
ENST00000430420.5 ENST00000453081.5 ENST00000319167.12 ENST00000622017.4 ENST00000375022.8 ENST00000399736.5 ENST00000440072.5 ENST00000173898.12 ENST00000431115.5 ENST00000440759.5 ENST00000375041.6 |
TRO
|
trophinin |
| chr6_+_106086316 | 0.63 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr19_-_14778552 | 0.63 |
ENST00000315576.8
|
ADGRE2
|
adhesion G protein-coupled receptor E2 |
| chr19_+_7030578 | 0.63 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr18_+_63777773 | 0.62 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr22_-_26565362 | 0.61 |
ENST00000398110.6
|
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr1_+_115029823 | 0.60 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr19_+_14941489 | 0.60 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor family 7 subfamily C member 2 |
| chr21_-_43075831 | 0.60 |
ENST00000398158.5
ENST00000398165.8 |
CBS
|
cystathionine beta-synthase |
| chr19_+_11089446 | 0.60 |
ENST00000557933.5
ENST00000455727.6 ENST00000535915.5 ENST00000545707.5 ENST00000558518.6 ENST00000558013.5 |
LDLR
|
low density lipoprotein receptor |
| chr4_+_108650585 | 0.60 |
ENST00000613215.4
ENST00000361564.9 |
OSTC
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr18_+_63587297 | 0.59 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr19_-_42528380 | 0.58 |
ENST00000403461.5
ENST00000352591.9 ENST00000358394.7 ENST00000403444.7 ENST00000161559.11 ENST00000599389.1 |
CEACAM1
|
CEA cell adhesion molecule 1 |
| chr11_+_62856072 | 0.58 |
ENST00000377890.6
ENST00000681467.1 ENST00000538084.2 ENST00000681569.1 ENST00000377891.6 ENST00000680725.1 ENST00000377889.6 |
SLC3A2
|
solute carrier family 3 member 2 |
| chr6_-_27912396 | 0.56 |
ENST00000303324.4
|
OR2B2
|
olfactory receptor family 2 subfamily B member 2 |
| chr9_+_72577369 | 0.56 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr19_+_41376692 | 0.56 |
ENST00000447302.6
ENST00000544232.5 ENST00000542945.5 ENST00000540732.3 |
TMEM91
ENSG00000255730.5
|
transmembrane protein 91 novel protein |
| chr19_-_48740573 | 0.56 |
ENST00000222145.9
|
RASIP1
|
Ras interacting protein 1 |
| chr3_+_26694499 | 0.56 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr11_+_89924064 | 0.55 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr21_-_43076362 | 0.55 |
ENST00000359624.7
ENST00000352178.9 |
CBS
|
cystathionine beta-synthase |
| chrX_-_66033664 | 0.55 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr2_-_88947820 | 0.55 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr2_-_186849164 | 0.55 |
ENST00000295131.3
|
ZSWIM2
|
zinc finger SWIM-type containing 2 |
| chr6_-_26285526 | 0.54 |
ENST00000377727.2
|
H4C8
|
H4 clustered histone 8 |
| chr4_+_70721953 | 0.53 |
ENST00000381006.8
ENST00000226328.8 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr22_-_50085331 | 0.53 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr1_-_53838276 | 0.53 |
ENST00000371429.4
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr11_+_94768331 | 0.53 |
ENST00000317829.12
ENST00000433060.3 |
AMOTL1
|
angiomotin like 1 |
| chr7_-_36724543 | 0.53 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr11_-_48983826 | 0.52 |
ENST00000649162.1
|
TRIM51GP
|
tripartite motif-containing 51G, pseudogene |
| chr3_+_130931893 | 0.52 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr4_+_186266183 | 0.52 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr7_-_36724457 | 0.51 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr6_+_31137646 | 0.51 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr1_+_155209213 | 0.51 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr22_-_50085414 | 0.51 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chrX_-_47629845 | 0.51 |
ENST00000469388.1
ENST00000396992.8 ENST00000377005.6 |
CFP
|
complement factor properdin |
| chr9_+_107306459 | 0.50 |
ENST00000457811.1
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr6_+_29100609 | 0.50 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr3_+_44874606 | 0.50 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr4_-_142305935 | 0.50 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr14_+_24070837 | 0.49 |
ENST00000537691.5
ENST00000397016.6 ENST00000560356.5 ENST00000558450.5 |
CPNE6
|
copine 6 |
| chr19_-_58353482 | 0.49 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_-_19060706 | 0.49 |
ENST00000329773.3
|
MRGPRX2
|
MAS related GPR family member X2 |
| chr7_+_142750657 | 0.49 |
ENST00000492062.1
|
PRSS1
|
serine protease 1 |
| chr19_-_52171643 | 0.49 |
ENST00000597065.1
|
ZNF836
|
zinc finger protein 836 |
| chrX_+_86714623 | 0.49 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr3_+_136957948 | 0.49 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr8_+_81280527 | 0.49 |
ENST00000297258.11
|
FABP5
|
fatty acid binding protein 5 |
| chr8_-_143953845 | 0.48 |
ENST00000354958.6
|
PLEC
|
plectin |
| chr1_+_86993009 | 0.48 |
ENST00000370548.3
|
ENSG00000267561.2
|
novel protein |
| chr4_+_108650644 | 0.47 |
ENST00000512478.2
|
OSTC
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr8_-_123025750 | 0.47 |
ENST00000523036.1
|
DERL1
|
derlin 1 |
| chr3_-_197226351 | 0.47 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr4_-_69961007 | 0.46 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr7_-_99679987 | 0.46 |
ENST00000222982.8
ENST00000439761.3 ENST00000339843.6 |
CYP3A5
|
cytochrome P450 family 3 subfamily A member 5 |
| chr2_-_171434763 | 0.46 |
ENST00000442778.5
ENST00000453846.5 ENST00000612742.5 |
METTL8
|
methyltransferase like 8 |
| chr8_+_18391276 | 0.46 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr16_-_18562074 | 0.45 |
ENST00000543392.5
ENST00000622306.5 ENST00000381474.7 ENST00000621364.4 ENST00000330537.10 |
NOMO2
|
NODAL modulator 2 |
| chr5_+_168529299 | 0.45 |
ENST00000338333.5
|
FBLL1
|
fibrillarin like 1 |
| chr1_+_56854764 | 0.45 |
ENST00000361249.4
|
C8A
|
complement C8 alpha chain |
| chr19_-_42427379 | 0.45 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr16_+_22505845 | 0.45 |
ENST00000356156.7
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr4_-_151227881 | 0.44 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr4_-_159035226 | 0.44 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr8_-_123025627 | 0.44 |
ENST00000519018.5
|
DERL1
|
derlin 1 |
| chr20_-_22585451 | 0.44 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr1_+_196819731 | 0.43 |
ENST00000320493.10
ENST00000367424.4 |
CFHR1
|
complement factor H related 1 |
| chr3_+_197950176 | 0.42 |
ENST00000448864.6
ENST00000647248.2 |
RPL35A
|
ribosomal protein L35a |
| chr3_+_102099244 | 0.42 |
ENST00000491959.5
|
ZPLD1
|
zona pellucida like domain containing 1 |
| chr17_-_82840010 | 0.42 |
ENST00000269394.4
ENST00000572562.1 |
ZNF750
|
zinc finger protein 750 |
| chr3_+_159839847 | 0.42 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr3_+_35641421 | 0.41 |
ENST00000449196.5
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr4_+_73409340 | 0.41 |
ENST00000511370.1
|
ALB
|
albumin |
| chr11_-_55936400 | 0.40 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr5_-_16742221 | 0.40 |
ENST00000505695.5
|
MYO10
|
myosin X |
| chr7_+_151956379 | 0.40 |
ENST00000431418.6
|
GALNTL5
|
polypeptide N-acetylgalactosaminyltransferase like 5 |
| chr8_+_12104389 | 0.39 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr3_+_191329020 | 0.39 |
ENST00000392456.4
|
CCDC50
|
coiled-coil domain containing 50 |
| chr21_+_38272291 | 0.39 |
ENST00000438657.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr17_-_66229380 | 0.39 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr4_+_176319942 | 0.39 |
ENST00000503362.2
|
SPCS3
|
signal peptidase complex subunit 3 |
| chr12_-_64390727 | 0.39 |
ENST00000543942.7
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr18_-_5396265 | 0.38 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr9_-_21187671 | 0.38 |
ENST00000421715.2
|
IFNA4
|
interferon alpha 4 |
| chr1_-_75932392 | 0.38 |
ENST00000284142.7
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr5_-_156963222 | 0.38 |
ENST00000407087.4
ENST00000274532.7 |
TIMD4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr21_+_38272410 | 0.38 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr16_-_69754913 | 0.38 |
ENST00000268802.10
|
NOB1
|
NIN1 (RPN12) binding protein 1 homolog |
| chr10_+_49637400 | 0.37 |
ENST00000640822.1
|
CHAT
|
choline O-acetyltransferase |
| chr15_-_65133780 | 0.37 |
ENST00000204549.9
|
PDCD7
|
programmed cell death 7 |
| chr13_+_57140918 | 0.37 |
ENST00000377931.1
ENST00000614894.4 |
PRR20A
PRR20C
|
proline rich 20A proline rich 20C |
| chr12_-_10453330 | 0.37 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr11_+_57597563 | 0.37 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr11_+_56176618 | 0.37 |
ENST00000312298.1
|
OR5J2
|
olfactory receptor family 5 subfamily J member 2 |
| chr12_-_51324652 | 0.36 |
ENST00000544402.5
|
BIN2
|
bridging integrator 2 |
| chr5_+_58491427 | 0.36 |
ENST00000396776.6
ENST00000502276.6 ENST00000511930.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr3_-_173141227 | 0.36 |
ENST00000351008.4
|
SPATA16
|
spermatogenesis associated 16 |
| chr12_+_80099535 | 0.36 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr5_+_58491451 | 0.36 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr1_+_196774813 | 0.36 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr14_+_21990357 | 0.36 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr2_+_33436304 | 0.35 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr19_-_4400418 | 0.35 |
ENST00000598564.5
ENST00000417295.6 ENST00000269886.7 |
SH3GL1
|
SH3 domain containing GRB2 like 1, endophilin A2 |
| chr8_-_109974688 | 0.35 |
ENST00000297404.1
|
KCNV1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr19_+_3224701 | 0.35 |
ENST00000541430.6
|
CELF5
|
CUGBP Elav-like family member 5 |
| chr1_+_77779618 | 0.35 |
ENST00000370791.7
ENST00000443751.3 ENST00000645756.1 ENST00000643390.1 ENST00000642959.1 |
MIGA1
|
mitoguardin 1 |
| chr6_-_138218491 | 0.35 |
ENST00000527246.3
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr17_+_58238426 | 0.35 |
ENST00000421678.6
ENST00000262290.9 ENST00000543544.5 |
LPO
|
lactoperoxidase |
| chr12_-_10909562 | 0.34 |
ENST00000390677.2
|
TAS2R13
|
taste 2 receptor member 13 |
| chr4_+_15339818 | 0.34 |
ENST00000397700.6
ENST00000295297.4 |
C1QTNF7
|
C1q and TNF related 7 |
| chr5_+_127649018 | 0.34 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr12_+_112906777 | 0.34 |
ENST00000452357.7
ENST00000445409.7 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr11_+_65890627 | 0.33 |
ENST00000312579.4
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr1_-_247536440 | 0.33 |
ENST00000366487.4
ENST00000641802.1 |
OR2C3
|
olfactory receptor family 2 subfamily C member 3 |
| chr6_-_116829037 | 0.33 |
ENST00000368549.7
ENST00000530250.1 ENST00000310357.8 |
GPRC6A
|
G protein-coupled receptor class C group 6 member A |
| chr3_+_158110052 | 0.33 |
ENST00000295930.7
ENST00000471994.5 ENST00000482822.3 ENST00000476899.6 ENST00000683899.1 ENST00000684604.1 ENST00000682164.1 ENST00000464171.5 ENST00000611884.5 ENST00000312179.10 ENST00000475278.6 |
RSRC1
|
arginine and serine rich coiled-coil 1 |
| chr14_-_91732059 | 0.33 |
ENST00000553329.5
ENST00000256343.8 |
CATSPERB
|
cation channel sperm associated auxiliary subunit beta |
| chr9_+_122370523 | 0.33 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr4_-_68951763 | 0.32 |
ENST00000251566.9
|
UGT2A3
|
UDP glucuronosyltransferase family 2 member A3 |
| chr9_+_94084458 | 0.32 |
ENST00000620992.5
ENST00000288976.3 |
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr12_+_75334655 | 0.32 |
ENST00000378695.9
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr20_-_58228653 | 0.32 |
ENST00000457363.1
|
ANKRD60
|
ankyrin repeat domain 60 |
| chr2_+_113406368 | 0.32 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr7_+_57450171 | 0.32 |
ENST00000420713.2
|
ZNF716
|
zinc finger protein 716 |
| chr4_-_89835617 | 0.31 |
ENST00000611107.1
ENST00000345009.8 ENST00000505199.5 ENST00000502987.5 |
SNCA
|
synuclein alpha |
| chr6_-_75284820 | 0.31 |
ENST00000475111.6
|
TMEM30A
|
transmembrane protein 30A |
| chr7_+_151956440 | 0.31 |
ENST00000392800.7
ENST00000616416.4 |
GALNTL5
|
polypeptide N-acetylgalactosaminyltransferase like 5 |
| chrY_-_24047969 | 0.31 |
ENST00000306882.4
ENST00000382407.1 |
CDY1B
|
chromodomain Y-linked 1B |
| chr6_-_26216673 | 0.31 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chr3_+_8501807 | 0.31 |
ENST00000426878.2
ENST00000397386.7 ENST00000415597.5 ENST00000157600.8 |
LMCD1
|
LIM and cysteine rich domains 1 |
| chr4_+_69931066 | 0.31 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr11_+_44095710 | 0.31 |
ENST00000358681.8
ENST00000343631.4 ENST00000682711.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr11_+_122838492 | 0.31 |
ENST00000227348.9
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr11_+_7088991 | 0.31 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr6_-_29087313 | 0.31 |
ENST00000377173.4
|
OR2B3
|
olfactory receptor family 2 subfamily B member 3 |
| chr2_+_100974849 | 0.31 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr12_+_8989612 | 0.30 |
ENST00000266551.8
|
KLRG1
|
killer cell lectin like receptor G1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.4 | 1.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.4 | 3.2 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.6 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.6 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 0.8 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 0.6 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 1.2 | GO:0046439 | cysteine biosynthetic process from serine(GO:0006535) cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) homocysteine catabolic process(GO:0043418) L-cysteine metabolic process(GO:0046439) |
| 0.2 | 1.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 3.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 0.5 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 0.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 1.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.5 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.2 | 2.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 0.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.8 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 0.5 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 0.9 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.2 | 0.5 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.6 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.4 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.4 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 1.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.4 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.4 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 1.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.5 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 2.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.3 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.1 | 1.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.1 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.9 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.6 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 1.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 1.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 1.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 2.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.2 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.2 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 1.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.3 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.5 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 1.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.1 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 1.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.5 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.4 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 1.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 4.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.2 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 2.1 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.9 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:1905244 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) skeletal muscle satellite cell migration(GO:1902766) regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 2.2 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.3 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.1 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.4 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 1.1 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 1.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.5 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.5 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 1.3 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.3 | 1.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 0.6 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 1.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.9 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.3 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 3.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.1 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 1.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.7 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 2.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.7 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.4 | 2.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 3.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 1.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.3 | 1.2 | GO:0050421 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.3 | 0.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 1.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 0.5 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 2.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 2.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.3 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.4 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 1.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.2 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 2.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 1.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 3.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 3.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 3.4 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 4.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.7 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 1.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |