Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for DBX2_HLX
Z-value: 0.45
Transcription factors associated with DBX2_HLX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
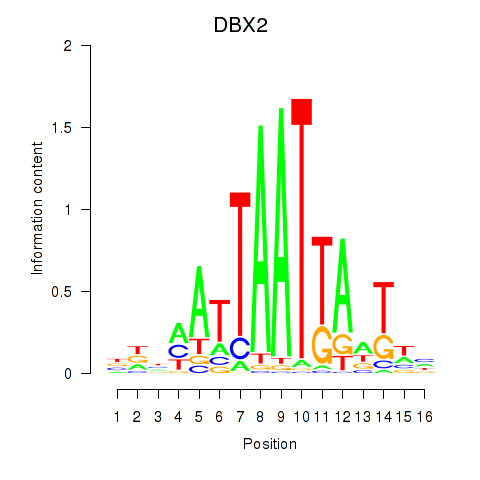
DBX2
|
ENSG00000185610.6 | DBX2 |
|
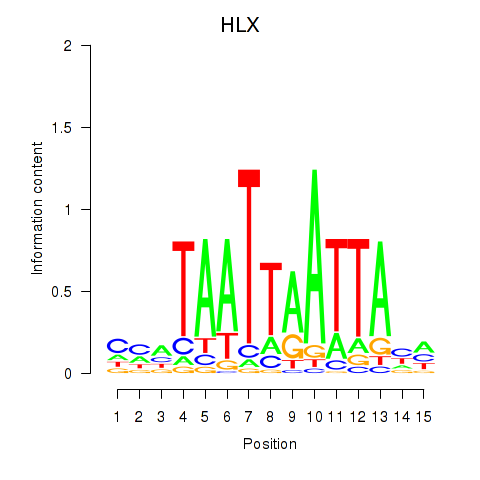
HLX
|
ENSG00000136630.13 | HLX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLX | hg38_v1_chr1_+_220879434_220879457 | 0.09 | 6.4e-01 | Click! |
Activity profile of DBX2_HLX motif
Sorted Z-values of DBX2_HLX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DBX2_HLX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_197067234 | 1.09 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr3_-_142029108 | 1.08 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr9_+_12693327 | 1.06 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr6_-_25874212 | 0.91 |
ENST00000361703.10
ENST00000397060.8 |
SLC17A3
|
solute carrier family 17 member 3 |
| chr7_+_123601815 | 0.91 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_123601836 | 0.82 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr6_-_27912396 | 0.80 |
ENST00000303324.4
|
OR2B2
|
olfactory receptor family 2 subfamily B member 2 |
| chr8_-_85341705 | 0.70 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr3_-_194351290 | 0.66 |
ENST00000429275.1
ENST00000323830.4 |
CPN2
|
carboxypeptidase N subunit 2 |
| chr20_-_7940444 | 0.63 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr11_-_5301946 | 0.62 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr5_-_147401591 | 0.57 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr4_-_149815826 | 0.57 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr2_+_102337148 | 0.57 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr4_-_159035226 | 0.56 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr15_-_19988117 | 0.56 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr12_-_10826358 | 0.53 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr9_+_27109393 | 0.45 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr19_+_53962925 | 0.44 |
ENST00000270458.4
|
CACNG8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr12_-_8066331 | 0.43 |
ENST00000546241.1
ENST00000307637.5 |
C3AR1
|
complement C3a receptor 1 |
| chr12_-_91111460 | 0.43 |
ENST00000266718.5
|
LUM
|
lumican |
| chr21_-_30497160 | 0.43 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr5_-_140346596 | 0.43 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr3_-_33645433 | 0.43 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_+_35180279 | 0.41 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr9_-_21482313 | 0.40 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr4_+_155666718 | 0.40 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr19_+_926001 | 0.39 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr16_+_56191476 | 0.39 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr3_+_133124807 | 0.39 |
ENST00000508711.5
|
TMEM108
|
transmembrane protein 108 |
| chr5_-_126595237 | 0.38 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr1_+_76867469 | 0.38 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr10_+_89392546 | 0.38 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chrX_-_15314543 | 0.36 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_-_247758680 | 0.36 |
ENST00000408896.4
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr21_+_32298945 | 0.36 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr12_-_84892120 | 0.34 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr6_+_123804141 | 0.34 |
ENST00000368416.5
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr7_+_142111739 | 0.34 |
ENST00000550469.6
ENST00000477922.3 |
MGAM2
|
maltase-glucoamylase 2 (putative) |
| chr10_+_96000091 | 0.34 |
ENST00000424464.5
ENST00000410012.6 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr1_+_76074698 | 0.33 |
ENST00000328299.4
|
ST6GALNAC3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr6_+_85449584 | 0.33 |
ENST00000369651.7
|
NT5E
|
5'-nucleotidase ecto |
| chr11_-_122116215 | 0.33 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr11_-_107858777 | 0.33 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr19_-_3557563 | 0.33 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr17_-_41149823 | 0.32 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr7_-_93890160 | 0.31 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr1_+_86468902 | 0.31 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr3_-_48089203 | 0.31 |
ENST00000468075.2
ENST00000360240.10 |
MAP4
|
microtubule associated protein 4 |
| chr2_+_54558348 | 0.31 |
ENST00000333896.5
|
SPTBN1
|
spectrin beta, non-erythrocytic 1 |
| chr19_+_9185594 | 0.30 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr15_-_79971164 | 0.30 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr3_-_112845950 | 0.29 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr11_-_7796942 | 0.29 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2 |
| chr20_+_3786772 | 0.29 |
ENST00000344256.10
ENST00000379598.9 |
CDC25B
|
cell division cycle 25B |
| chr9_-_21305313 | 0.29 |
ENST00000610521.2
|
IFNA5
|
interferon alpha 5 |
| chr7_-_122699108 | 0.29 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr10_+_127998126 | 0.27 |
ENST00000455661.5
|
PTPRE
|
protein tyrosine phosphatase receptor type E |
| chr12_-_95116967 | 0.27 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr9_+_36572854 | 0.27 |
ENST00000543751.5
ENST00000536860.5 ENST00000541717.4 ENST00000536329.5 ENST00000536987.5 ENST00000298048.7 ENST00000495529.5 ENST00000545008.5 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr19_-_42877988 | 0.27 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr2_-_224947030 | 0.27 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr15_+_76336755 | 0.26 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr6_-_76072654 | 0.26 |
ENST00000369950.8
ENST00000611179.4 ENST00000369963.5 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr19_+_16829387 | 0.26 |
ENST00000248054.10
ENST00000596802.5 ENST00000379803.5 |
SIN3B
|
SIN3 transcription regulator family member B |
| chr13_+_31739542 | 0.25 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chrX_+_136205982 | 0.25 |
ENST00000628568.1
|
FHL1
|
four and a half LIM domains 1 |
| chr4_-_109801978 | 0.25 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr12_-_7503841 | 0.25 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr4_+_94207596 | 0.25 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_-_35265696 | 0.25 |
ENST00000464522.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr16_+_81645352 | 0.25 |
ENST00000398040.8
|
CMIP
|
c-Maf inducing protein |
| chr6_+_130018565 | 0.25 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr2_+_186694007 | 0.24 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr7_+_138076453 | 0.24 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr2_+_89936859 | 0.24 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr2_+_90172802 | 0.24 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr7_+_134891566 | 0.24 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr1_+_212035717 | 0.23 |
ENST00000366991.5
|
DTL
|
denticleless E3 ubiquitin protein ligase homolog |
| chr22_-_28712136 | 0.23 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2 |
| chr11_-_123654939 | 0.23 |
ENST00000657191.1
|
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr8_-_13276491 | 0.23 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr13_+_31739520 | 0.23 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr4_-_137532452 | 0.23 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr2_+_102418642 | 0.23 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr17_-_445939 | 0.22 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr9_+_74615582 | 0.22 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr19_-_4535221 | 0.22 |
ENST00000381848.7
ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr10_+_52128343 | 0.21 |
ENST00000672084.1
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr2_+_181985846 | 0.21 |
ENST00000682840.1
ENST00000409137.7 ENST00000280295.7 |
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C |
| chr4_-_122621011 | 0.21 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr1_-_247760556 | 0.21 |
ENST00000641256.1
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr16_+_11965234 | 0.21 |
ENST00000562385.1
|
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr6_-_87095059 | 0.21 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr2_-_189179754 | 0.21 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr1_+_244352627 | 0.21 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr11_-_60183191 | 0.21 |
ENST00000412309.6
|
MS4A6A
|
membrane spanning 4-domains A6A |
| chr5_-_22853320 | 0.21 |
ENST00000504376.6
ENST00000382254.6 |
CDH12
|
cadherin 12 |
| chr2_+_29097705 | 0.21 |
ENST00000401605.5
ENST00000401617.6 |
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr5_-_112419251 | 0.21 |
ENST00000261486.6
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr15_+_78565556 | 0.21 |
ENST00000559554.5
|
CHRNA5
|
cholinergic receptor nicotinic alpha 5 subunit |
| chr7_+_130486171 | 0.21 |
ENST00000341441.9
ENST00000416162.7 |
MEST
|
mesoderm specific transcript |
| chrX_+_15507302 | 0.21 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chrX_+_83508284 | 0.20 |
ENST00000644024.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr9_-_92404559 | 0.20 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr16_+_11345429 | 0.20 |
ENST00000576027.1
ENST00000312499.6 ENST00000648619.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr3_-_150703943 | 0.20 |
ENST00000491361.5
|
ERICH6
|
glutamate rich 6 |
| chr6_-_110179995 | 0.20 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr16_-_20404731 | 0.20 |
ENST00000302451.9
|
PDILT
|
protein disulfide isomerase like, testis expressed |
| chr9_-_39239174 | 0.20 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr4_+_70050431 | 0.20 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
| chr12_-_11395556 | 0.20 |
ENST00000565533.1
ENST00000389362.6 ENST00000546254.3 |
PRB2
PRB1
|
proline rich protein BstNI subfamily 2 proline rich protein BstNI subfamily 1 |
| chr11_-_117316230 | 0.20 |
ENST00000313005.11
ENST00000528053.5 |
BACE1
|
beta-secretase 1 |
| chr7_-_36985060 | 0.20 |
ENST00000396040.6
|
ELMO1
|
engulfment and cell motility 1 |
| chr7_+_130486324 | 0.20 |
ENST00000427521.6
ENST00000378576.9 |
MEST
|
mesoderm specific transcript |
| chr2_-_157444044 | 0.19 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr5_+_55160161 | 0.19 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr3_+_102099244 | 0.19 |
ENST00000491959.5
|
ZPLD1
|
zona pellucida like domain containing 1 |
| chr15_+_67125707 | 0.19 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr6_+_29111560 | 0.19 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr3_+_172754457 | 0.19 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr11_+_49028823 | 0.19 |
ENST00000332682.9
|
TRIM49B
|
tripartite motif containing 49B |
| chr14_-_67412112 | 0.18 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr10_-_5404819 | 0.18 |
ENST00000380419.8
ENST00000479328.1 |
TUBAL3
|
tubulin alpha like 3 |
| chr9_+_27109135 | 0.18 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr3_-_151203201 | 0.18 |
ENST00000480322.1
ENST00000309180.6 |
GPR171
|
G protein-coupled receptor 171 |
| chr15_+_78565507 | 0.18 |
ENST00000299565.9
|
CHRNA5
|
cholinergic receptor nicotinic alpha 5 subunit |
| chr12_+_75480745 | 0.18 |
ENST00000266659.8
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr17_+_18744026 | 0.18 |
ENST00000308799.8
ENST00000395665.9 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr15_+_21579912 | 0.18 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr12_+_75480800 | 0.18 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr8_-_48921419 | 0.17 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr10_+_60778331 | 0.17 |
ENST00000519078.6
ENST00000316629.8 ENST00000395284.8 |
CDK1
|
cyclin dependent kinase 1 |
| chr1_+_152811971 | 0.17 |
ENST00000360090.4
|
LCE1B
|
late cornified envelope 1B |
| chr6_-_107824294 | 0.17 |
ENST00000369020.8
ENST00000369022.6 |
SCML4
|
Scm polycomb group protein like 4 |
| chr12_+_26011713 | 0.17 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr9_-_21368962 | 0.17 |
ENST00000610660.1
|
IFNA13
|
interferon alpha 13 |
| chr4_-_151227881 | 0.17 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr13_-_46105009 | 0.17 |
ENST00000439329.5
ENST00000674625.1 ENST00000181383.10 |
CPB2
|
carboxypeptidase B2 |
| chr11_+_121102666 | 0.17 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr17_-_3127551 | 0.17 |
ENST00000328890.3
|
OR1G1
|
olfactory receptor family 1 subfamily G member 1 |
| chr15_+_41286011 | 0.17 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr9_+_27109200 | 0.17 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr1_+_196888014 | 0.17 |
ENST00000367416.6
ENST00000608469.6 ENST00000251424.8 ENST00000367418.2 |
CFHR4
|
complement factor H related 4 |
| chr10_+_69088096 | 0.17 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr7_+_142800957 | 0.17 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr12_-_86256267 | 0.16 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr2_+_190137760 | 0.16 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr12_+_75481204 | 0.16 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr1_+_84164962 | 0.16 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr12_+_8822610 | 0.16 |
ENST00000299698.12
|
A2ML1
|
alpha-2-macroglobulin like 1 |
| chr3_+_44874606 | 0.16 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr9_-_21217311 | 0.16 |
ENST00000380216.1
|
IFNA16
|
interferon alpha 16 |
| chr1_-_153150884 | 0.16 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr21_-_30813270 | 0.15 |
ENST00000329621.6
|
KRTAP8-1
|
keratin associated protein 8-1 |
| chr1_+_149782671 | 0.15 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chr17_+_43847142 | 0.15 |
ENST00000377203.8
ENST00000539718.5 ENST00000588884.1 ENST00000293396.12 ENST00000586233.5 |
CD300LG
|
CD300 molecule like family member g |
| chr6_+_29301701 | 0.15 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr22_+_44752552 | 0.15 |
ENST00000389774.6
ENST00000356099.11 ENST00000396119.6 ENST00000336963.8 ENST00000412433.5 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr14_-_106593319 | 0.15 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr2_-_162838728 | 0.15 |
ENST00000328032.8
ENST00000332142.10 |
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr8_+_12104389 | 0.15 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr13_+_76948500 | 0.15 |
ENST00000377462.6
|
ACOD1
|
aconitate decarboxylase 1 |
| chr11_-_35360050 | 0.15 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr14_-_70080180 | 0.15 |
ENST00000394330.6
ENST00000533541.1 ENST00000216568.11 |
SLC8A3
|
solute carrier family 8 member A3 |
| chr17_-_40665121 | 0.15 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr7_+_134891400 | 0.15 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr5_-_88823763 | 0.14 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr11_+_57597563 | 0.14 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr6_+_34757473 | 0.14 |
ENST00000244520.10
ENST00000374018.5 ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr3_-_112829367 | 0.14 |
ENST00000448932.4
ENST00000617549.3 |
CD200R1L
|
CD200 receptor 1 like |
| chr6_+_154039333 | 0.14 |
ENST00000428397.6
|
OPRM1
|
opioid receptor mu 1 |
| chr10_-_114144599 | 0.14 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr19_-_42427379 | 0.14 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr5_+_151020438 | 0.14 |
ENST00000622181.4
ENST00000614343.4 ENST00000388825.9 ENST00000521650.5 ENST00000517973.1 |
GPX3
|
glutathione peroxidase 3 |
| chr10_+_100462969 | 0.14 |
ENST00000343737.6
|
WNT8B
|
Wnt family member 8B |
| chr21_+_14216145 | 0.13 |
ENST00000400577.4
|
RBM11
|
RNA binding motif protein 11 |
| chr8_-_113377125 | 0.13 |
ENST00000343508.7
|
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr10_-_13972355 | 0.13 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr2_-_177888728 | 0.13 |
ENST00000389683.7
|
PDE11A
|
phosphodiesterase 11A |
| chr3_+_63443076 | 0.13 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr17_+_80544817 | 0.13 |
ENST00000306801.8
ENST00000570891.5 |
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr19_-_51417791 | 0.13 |
ENST00000353836.9
|
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr14_+_103385450 | 0.13 |
ENST00000416682.6
|
MARK3
|
microtubule affinity regulating kinase 3 |
| chr4_+_87832917 | 0.13 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chrX_+_154304923 | 0.13 |
ENST00000426989.5
ENST00000426203.5 ENST00000369912.2 |
TKTL1
|
transketolase like 1 |
| chr2_+_90038848 | 0.13 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr4_-_162163955 | 0.13 |
ENST00000379164.8
|
FSTL5
|
follistatin like 5 |
| chr4_-_76036060 | 0.13 |
ENST00000306621.8
|
CXCL11
|
C-X-C motif chemokine ligand 11 |
| chr19_-_51417700 | 0.12 |
ENST00000529627.1
ENST00000439889.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr4_+_70383123 | 0.12 |
ENST00000304915.8
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr16_-_28211476 | 0.12 |
ENST00000569951.1
ENST00000565698.5 |
XPO6
|
exportin 6 |
| chr18_+_45615473 | 0.12 |
ENST00000255226.11
|
SLC14A2
|
solute carrier family 14 member 2 |
| chr6_+_46793379 | 0.12 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr10_+_89301932 | 0.12 |
ENST00000371826.4
ENST00000679755.1 |
IFIT2
|
interferon induced protein with tetratricopeptide repeats 2 |
| chr15_+_21651844 | 0.12 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr14_+_21868822 | 0.12 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr11_+_4329865 | 0.12 |
ENST00000640302.1
|
SSU72P3
|
SSU72 pseudogene 3 |
| chr17_+_29941605 | 0.12 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr8_-_61646807 | 0.12 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.3 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.6 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.2 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.4 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.5 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0071393 | propionate metabolic process(GO:0019541) cellular response to progesterone stimulus(GO:0071393) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.0 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.2 | GO:0090166 | regulation of Schwann cell differentiation(GO:0014038) Golgi disassembly(GO:0090166) |
| 0.0 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 2.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 1.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.0 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.0 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030849 | X chromosome(GO:0000805) autosome(GO:0030849) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 1.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0071753 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.2 | 0.6 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.9 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 0.3 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.3 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |