Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for DPRX
Z-value: 0.52
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | DPRX |
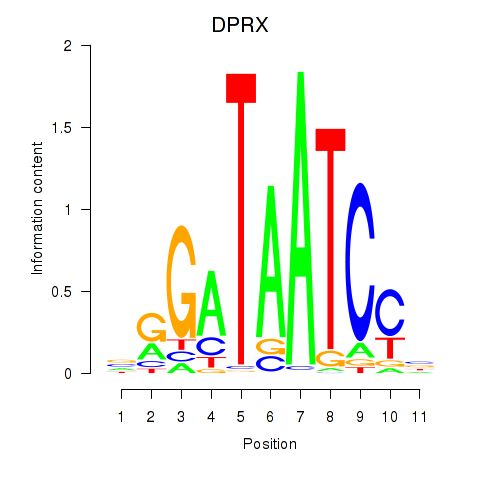
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_123601815 | 1.10 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_123601836 | 1.10 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr19_+_7030578 | 0.65 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr19_-_7021431 | 0.55 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr9_+_98943705 | 0.53 |
ENST00000610452.1
|
COL15A1
|
collagen type XV alpha 1 chain |
| chr9_-_13279407 | 0.52 |
ENST00000546205.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr19_-_7040179 | 0.52 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4 |
| chr22_-_30471986 | 0.51 |
ENST00000401751.5
ENST00000402286.5 ENST00000403066.5 ENST00000215812.9 |
SEC14L3
|
SEC14 like lipid binding 3 |
| chr19_+_7049321 | 0.49 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3 like 2 |
| chr16_-_28403841 | 0.46 |
ENST00000398944.7
ENST00000380876.5 |
EIF3CL
|
eukaryotic translation initiation factor 3 subunit C like |
| chr15_+_33310946 | 0.44 |
ENST00000415757.7
ENST00000634891.2 ENST00000389232.9 ENST00000622037.1 |
RYR3
|
ryanodine receptor 3 |
| chr7_+_150567382 | 0.43 |
ENST00000255945.4
ENST00000479232.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chrX_+_15507302 | 0.43 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr1_+_179591876 | 0.42 |
ENST00000294848.12
|
TDRD5
|
tudor domain containing 5 |
| chr1_+_179591819 | 0.42 |
ENST00000444136.6
|
TDRD5
|
tudor domain containing 5 |
| chr20_+_35172046 | 0.41 |
ENST00000216968.5
|
PROCR
|
protein C receptor |
| chr7_+_150567347 | 0.41 |
ENST00000461940.5
|
GIMAP4
|
GTPase, IMAP family member 4 |
| chr1_-_153549120 | 0.41 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_+_28412511 | 0.40 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr3_-_112845950 | 0.38 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr6_-_32763522 | 0.35 |
ENST00000435145.6
ENST00000437316.7 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr8_+_53851786 | 0.34 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr16_-_30011391 | 0.33 |
ENST00000616445.4
ENST00000564944.5 |
DOC2A
|
double C2 domain alpha |
| chr3_+_45943455 | 0.32 |
ENST00000304552.5
|
CXCR6
|
C-X-C motif chemokine receptor 6 |
| chrX_+_53422890 | 0.32 |
ENST00000457095.5
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr6_-_32816910 | 0.32 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr16_+_28711417 | 0.31 |
ENST00000395587.5
ENST00000569690.5 ENST00000331666.11 ENST00000564243.5 ENST00000566866.5 |
EIF3C
|
eukaryotic translation initiation factor 3 subunit C |
| chr6_-_37257622 | 0.30 |
ENST00000650812.1
ENST00000497775.1 ENST00000478262.2 ENST00000356757.7 |
ENSG00000286105.1
TMEM217
|
novel transmembrane protein transmembrane protein 217 |
| chr17_+_7579491 | 0.30 |
ENST00000380498.10
ENST00000584502.1 ENST00000250092.11 |
CD68
|
CD68 molecule |
| chr3_+_159273235 | 0.30 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr6_-_37257590 | 0.29 |
ENST00000336655.7
|
TMEM217
|
transmembrane protein 217 |
| chr6_-_37257643 | 0.29 |
ENST00000651039.1
ENST00000652495.1 ENST00000652218.1 |
TMEM217
|
transmembrane protein 217 |
| chr3_+_148730100 | 0.29 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr15_+_41286011 | 0.29 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr1_-_120054225 | 0.28 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr4_+_68447453 | 0.28 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr1_+_13171848 | 0.28 |
ENST00000415919.3
|
PRAMEF9
|
PRAME family member 9 |
| chr1_+_15410168 | 0.27 |
ENST00000445566.1
|
EFHD2
|
EF-hand domain family member D2 |
| chr1_-_54623518 | 0.27 |
ENST00000302250.7
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151 member A |
| chr2_-_49154433 | 0.26 |
ENST00000454032.5
ENST00000304421.8 |
FSHR
|
follicle stimulating hormone receptor |
| chr21_-_26845402 | 0.26 |
ENST00000284984.8
ENST00000676955.1 |
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr2_-_49154507 | 0.26 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor |
| chr11_-_66907891 | 0.25 |
ENST00000393955.6
|
PC
|
pyruvate carboxylase |
| chr9_-_13279564 | 0.25 |
ENST00000541718.5
ENST00000447879.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr12_-_52777343 | 0.24 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr1_-_153549238 | 0.24 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chrX_+_100644183 | 0.24 |
ENST00000640889.1
ENST00000373004.5 |
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr4_+_36281591 | 0.24 |
ENST00000639862.2
ENST00000357504.7 |
DTHD1
|
death domain containing 1 |
| chr6_-_22297028 | 0.22 |
ENST00000306482.2
|
PRL
|
prolactin |
| chr4_-_152779710 | 0.22 |
ENST00000304337.3
|
TIGD4
|
tigger transposable element derived 4 |
| chr18_+_44680875 | 0.22 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr12_-_70788914 | 0.22 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr14_+_70452161 | 0.21 |
ENST00000603540.2
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr1_-_13347134 | 0.21 |
ENST00000334600.7
|
PRAMEF14
|
PRAME family member 14 |
| chr14_+_22202561 | 0.21 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr11_-_35419098 | 0.20 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr11_-_35419213 | 0.20 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr3_+_98353854 | 0.20 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor family 5 subfamily K member 4 |
| chr7_+_135557904 | 0.20 |
ENST00000285968.11
|
NUP205
|
nucleoporin 205 |
| chr11_-_35419462 | 0.20 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr11_-_35418966 | 0.19 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr7_+_139829153 | 0.19 |
ENST00000652056.1
|
TBXAS1
|
thromboxane A synthase 1 |
| chr1_+_3454657 | 0.19 |
ENST00000378378.9
|
ARHGEF16
|
Rho guanine nucleotide exchange factor 16 |
| chr9_+_113444725 | 0.19 |
ENST00000374140.6
|
RGS3
|
regulator of G protein signaling 3 |
| chr9_+_37667997 | 0.19 |
ENST00000539465.5
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr11_-_124320197 | 0.18 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr18_+_34710307 | 0.18 |
ENST00000679796.1
|
DTNA
|
dystrobrevin alpha |
| chr22_-_28711931 | 0.18 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr5_-_181242236 | 0.18 |
ENST00000507756.5
|
RACK1
|
receptor for activated C kinase 1 |
| chr2_-_202870862 | 0.17 |
ENST00000454326.5
ENST00000432273.5 ENST00000450143.5 ENST00000411681.5 |
ICA1L
|
islet cell autoantigen 1 like |
| chr7_+_134779625 | 0.17 |
ENST00000454108.5
ENST00000361675.7 |
CALD1
|
caldesmon 1 |
| chr2_-_189179754 | 0.17 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr7_+_134779663 | 0.17 |
ENST00000361901.6
|
CALD1
|
caldesmon 1 |
| chr3_+_10026409 | 0.17 |
ENST00000287647.7
ENST00000676013.1 ENST00000675286.1 ENST00000419585.5 |
FANCD2
|
FA complementation group D2 |
| chr2_+_33436304 | 0.16 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr10_-_70602731 | 0.16 |
ENST00000441259.2
|
PRF1
|
perforin 1 |
| chr19_-_49867542 | 0.16 |
ENST00000600910.5
ENST00000322344.8 ENST00000600573.5 ENST00000596726.3 ENST00000638016.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr14_+_85533167 | 0.16 |
ENST00000682132.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr11_-_5269933 | 0.16 |
ENST00000396895.3
|
HBE1
|
hemoglobin subunit epsilon 1 |
| chr1_-_150808251 | 0.16 |
ENST00000271651.8
ENST00000676970.1 ENST00000679260.1 ENST00000676751.1 ENST00000677887.1 |
CTSK
|
cathepsin K |
| chr1_+_6555301 | 0.16 |
ENST00000333172.11
ENST00000351136.7 |
TAS1R1
|
taste 1 receptor member 1 |
| chr10_-_70602759 | 0.16 |
ENST00000373209.2
|
PRF1
|
perforin 1 |
| chr3_-_49021045 | 0.16 |
ENST00000440857.5
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr5_+_36166556 | 0.15 |
ENST00000677886.1
|
SKP2
|
S-phase kinase associated protein 2 |
| chr17_-_19867929 | 0.15 |
ENST00000361658.6
ENST00000395544.9 |
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr16_+_55479188 | 0.15 |
ENST00000219070.9
|
MMP2
|
matrix metallopeptidase 2 |
| chr7_-_30026617 | 0.15 |
ENST00000222803.10
|
FKBP14
|
FKBP prolyl isomerase 14 |
| chr8_+_8701891 | 0.15 |
ENST00000519106.2
|
CLDN23
|
claudin 23 |
| chr1_+_236394268 | 0.15 |
ENST00000334232.9
|
EDARADD
|
EDAR associated death domain |
| chr1_+_12773738 | 0.14 |
ENST00000357726.5
|
PRAMEF12
|
PRAME family member 12 |
| chr7_+_134891566 | 0.14 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr18_-_74457944 | 0.14 |
ENST00000400291.2
|
DIPK1C
|
divergent protein kinase domain 1C |
| chr1_+_240123121 | 0.14 |
ENST00000681210.1
|
FMN2
|
formin 2 |
| chr8_-_71356653 | 0.14 |
ENST00000388742.8
ENST00000388740.4 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr18_+_58862904 | 0.14 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532 |
| chr11_-_2301859 | 0.14 |
ENST00000456145.2
ENST00000381153.8 |
C11orf21
|
chromosome 11 open reading frame 21 |
| chr1_+_240123148 | 0.13 |
ENST00000681824.1
|
FMN2
|
formin 2 |
| chr16_-_15856994 | 0.13 |
ENST00000576790.7
ENST00000396324.7 ENST00000300036.6 ENST00000452625.7 |
MYH11
|
myosin heavy chain 11 |
| chr2_-_26478032 | 0.13 |
ENST00000338581.10
ENST00000402415.8 |
OTOF
|
otoferlin |
| chr13_+_51861963 | 0.13 |
ENST00000242819.7
|
CCDC70
|
coiled-coil domain containing 70 |
| chr2_+_233307806 | 0.12 |
ENST00000447536.5
ENST00000409110.6 |
SAG
|
S-antigen visual arrestin |
| chr12_+_53954870 | 0.12 |
ENST00000243103.4
|
HOXC12
|
homeobox C12 |
| chr15_+_22094522 | 0.12 |
ENST00000328795.5
|
OR4N4
|
olfactory receptor family 4 subfamily N member 4 |
| chr8_-_71356511 | 0.12 |
ENST00000419131.6
ENST00000388743.6 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_13201409 | 0.12 |
ENST00000625019.3
|
PRAMEF13
|
PRAME family member 13 |
| chr14_-_68979436 | 0.12 |
ENST00000193403.10
|
ACTN1
|
actinin alpha 1 |
| chr2_+_119759875 | 0.12 |
ENST00000263708.7
|
PTPN4
|
protein tyrosine phosphatase non-receptor type 4 |
| chr1_+_20070156 | 0.12 |
ENST00000375108.4
|
PLA2G5
|
phospholipase A2 group V |
| chr1_+_11691688 | 0.12 |
ENST00000294485.6
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr4_-_122621011 | 0.11 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr6_-_30690968 | 0.11 |
ENST00000376420.9
ENST00000376421.7 |
NRM
|
nurim |
| chr18_-_69947864 | 0.11 |
ENST00000582621.6
|
CD226
|
CD226 molecule |
| chr1_+_63773966 | 0.11 |
ENST00000371079.6
ENST00000371080.5 |
ROR1
|
receptor tyrosine kinase like orphan receptor 1 |
| chrX_-_103502853 | 0.11 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr5_-_135535955 | 0.11 |
ENST00000314744.6
|
NEUROG1
|
neurogenin 1 |
| chr2_-_26478113 | 0.11 |
ENST00000339598.8
|
OTOF
|
otoferlin |
| chr4_-_151227881 | 0.11 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr5_+_139342442 | 0.11 |
ENST00000394795.6
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chrX_+_77910656 | 0.11 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr2_+_68974573 | 0.10 |
ENST00000673932.3
ENST00000377938.4 |
GKN1
|
gastrokine 1 |
| chr6_+_46793379 | 0.10 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr4_-_56977577 | 0.10 |
ENST00000264230.5
|
NOA1
|
nitric oxide associated 1 |
| chr20_+_142573 | 0.10 |
ENST00000382398.4
|
DEFB126
|
defensin beta 126 |
| chr12_+_112907006 | 0.10 |
ENST00000680455.1
ENST00000551241.6 ENST00000550689.2 ENST00000679841.1 ENST00000679494.1 ENST00000553185.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr1_+_119507203 | 0.10 |
ENST00000369413.8
ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr3_+_49021071 | 0.10 |
ENST00000395458.6
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr6_+_157381133 | 0.10 |
ENST00000414563.6
ENST00000359775.10 |
ZDHHC14
|
zinc finger DHHC-type palmitoyltransferase 14 |
| chrX_-_72943837 | 0.10 |
ENST00000615063.2
|
DMRTC1
|
DMRT like family C1 |
| chr1_-_13155961 | 0.10 |
ENST00000624207.1
|
PRAMEF26
|
PRAME family member 26 |
| chr5_+_169583636 | 0.10 |
ENST00000506574.5
ENST00000515224.5 ENST00000629457.2 ENST00000508247.5 ENST00000265295.9 ENST00000513941.5 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr10_-_24723871 | 0.10 |
ENST00000396432.7
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr3_+_133038366 | 0.10 |
ENST00000321871.11
ENST00000393130.7 ENST00000514894.5 ENST00000512662.5 |
TMEM108
|
transmembrane protein 108 |
| chr6_-_31112566 | 0.09 |
ENST00000259870.4
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr1_-_173205543 | 0.09 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4 |
| chr8_-_30812867 | 0.09 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr15_+_21651844 | 0.09 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr11_+_65576023 | 0.09 |
ENST00000533237.5
ENST00000309295.9 ENST00000634639.1 |
EHBP1L1
|
EH domain binding protein 1 like 1 |
| chr1_-_9071716 | 0.09 |
ENST00000484798.5
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr1_-_101846957 | 0.09 |
ENST00000338858.9
|
OLFM3
|
olfactomedin 3 |
| chr15_+_81182579 | 0.09 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr12_+_112906777 | 0.09 |
ENST00000452357.7
ENST00000445409.7 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr7_+_30284574 | 0.09 |
ENST00000323037.5
|
ZNRF2
|
zinc and ring finger 2 |
| chr17_+_29593468 | 0.09 |
ENST00000614878.4
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr5_-_83720813 | 0.09 |
ENST00000515590.1
ENST00000274341.9 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr4_+_188002708 | 0.09 |
ENST00000618147.1
|
ZFP42
|
ZFP42 zinc finger protein |
| chr17_-_67996428 | 0.09 |
ENST00000580729.3
|
C17orf58
|
chromosome 17 open reading frame 58 |
| chr17_-_39225936 | 0.09 |
ENST00000333461.6
|
STAC2
|
SH3 and cysteine rich domain 2 |
| chr7_-_135728177 | 0.09 |
ENST00000682651.1
ENST00000354042.8 |
SLC13A4
|
solute carrier family 13 member 4 |
| chr5_+_139341875 | 0.08 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr1_+_224183262 | 0.08 |
ENST00000391877.3
|
DEGS1
|
delta 4-desaturase, sphingolipid 1 |
| chr11_-_85719111 | 0.08 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr5_+_139341826 | 0.08 |
ENST00000265192.9
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr16_+_51553436 | 0.08 |
ENST00000565308.2
|
HNRNPA1P48
|
heterogeneous nuclear ribonucleoprotein A1 pseudogene 48 |
| chr12_+_112906949 | 0.08 |
ENST00000679971.1
ENST00000675868.2 ENST00000550883.2 ENST00000553152.2 ENST00000202917.10 ENST00000679467.1 ENST00000680659.1 ENST00000540589.3 ENST00000552526.2 ENST00000681228.1 ENST00000680934.1 ENST00000681700.1 ENST00000679987.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr5_+_122129533 | 0.08 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr10_+_68901258 | 0.08 |
ENST00000373585.8
|
DDX50
|
DExD-box helicase 50 |
| chrX_+_15749848 | 0.08 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chrX_+_72776873 | 0.08 |
ENST00000334036.10
|
DMRTC1B
|
DMRT like family C1B |
| chr11_-_30586866 | 0.08 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr5_+_35856883 | 0.08 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr5_+_180909564 | 0.08 |
ENST00000505126.5
ENST00000533815.2 |
BTNL8
|
butyrophilin like 8 |
| chr11_-_85719160 | 0.08 |
ENST00000389958.7
ENST00000527794.5 |
SYTL2
|
synaptotagmin like 2 |
| chrX_-_139642889 | 0.08 |
ENST00000370576.9
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr14_+_75002903 | 0.08 |
ENST00000266126.10
|
EIF2B2
|
eukaryotic translation initiation factor 2B subunit beta |
| chr15_+_33968484 | 0.08 |
ENST00000383263.7
|
CHRM5
|
cholinergic receptor muscarinic 5 |
| chr3_-_42875871 | 0.08 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr18_+_34710249 | 0.08 |
ENST00000680346.1
ENST00000348997.9 ENST00000681274.1 ENST00000680822.1 ENST00000680767.2 ENST00000597599.5 ENST00000444659.6 |
DTNA
|
dystrobrevin alpha |
| chr7_-_98252117 | 0.07 |
ENST00000420697.1
ENST00000415086.5 ENST00000447648.7 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr17_+_29593118 | 0.07 |
ENST00000394859.8
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr6_-_127918540 | 0.07 |
ENST00000368250.5
|
THEMIS
|
thymocyte selection associated |
| chr1_-_170074568 | 0.07 |
ENST00000367767.5
ENST00000361580.7 ENST00000538366.5 |
KIFAP3
|
kinesin associated protein 3 |
| chrX_-_139642835 | 0.07 |
ENST00000536274.5
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr11_+_65181194 | 0.07 |
ENST00000533820.5
|
CAPN1
|
calpain 1 |
| chr18_-_66604076 | 0.07 |
ENST00000540086.5
ENST00000580157.2 ENST00000262150.7 |
CDH19
|
cadherin 19 |
| chr17_+_5420172 | 0.07 |
ENST00000381209.8
ENST00000405578.8 ENST00000574003.1 |
RPAIN
|
RPA interacting protein |
| chr18_-_3845292 | 0.07 |
ENST00000400145.6
|
DLGAP1
|
DLG associated protein 1 |
| chr7_-_155809072 | 0.07 |
ENST00000430104.5
|
SHH
|
sonic hedgehog signaling molecule |
| chr7_+_134891400 | 0.07 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr19_+_40717091 | 0.07 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr1_-_161044941 | 0.07 |
ENST00000368020.5
|
USF1
|
upstream transcription factor 1 |
| chr1_+_13070853 | 0.07 |
ENST00000619661.2
|
PRAMEF25
|
PRAME family member 25 |
| chr10_-_101818405 | 0.07 |
ENST00000357797.9
ENST00000370094.7 |
OGA
|
O-GlcNAcase |
| chr14_+_85530127 | 0.07 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr17_-_75667165 | 0.07 |
ENST00000584999.1
ENST00000420326.6 ENST00000340830.9 |
RECQL5
|
RecQ like helicase 5 |
| chr19_+_35358460 | 0.07 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3 |
| chr6_-_32763466 | 0.07 |
ENST00000427449.1
ENST00000411527.5 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr10_+_88586762 | 0.07 |
ENST00000371939.7
|
LIPJ
|
lipase family member J |
| chr1_+_159439722 | 0.07 |
ENST00000641630.1
ENST00000423932.6 |
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr9_-_92482499 | 0.07 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr10_+_15043937 | 0.07 |
ENST00000378228.8
ENST00000378217.3 |
OLAH
|
oleoyl-ACP hydrolase |
| chr1_+_207089233 | 0.07 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr9_-_14300231 | 0.07 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr2_-_215138603 | 0.06 |
ENST00000272895.12
|
ABCA12
|
ATP binding cassette subfamily A member 12 |
| chr1_+_207089283 | 0.06 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr11_-_85719045 | 0.06 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr1_+_207088825 | 0.06 |
ENST00000367078.8
|
C4BPB
|
complement component 4 binding protein beta |
| chr14_+_85530163 | 0.06 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr12_-_101830926 | 0.06 |
ENST00000299314.12
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta |
| chr10_+_95755652 | 0.06 |
ENST00000371207.8
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr17_-_49848017 | 0.06 |
ENST00000326219.5
ENST00000334568.8 ENST00000352793.6 ENST00000398154.5 ENST00000436235.5 |
TAC4
|
tachykinin precursor 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0007309 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.2 | 0.5 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.3 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.8 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.4 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0071626 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0060003 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.3 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.2 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.3 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0032445 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.5 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.8 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 0.4 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 1.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004618 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.3 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0016297 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |