Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for DRGX_PROP1
Z-value: 0.92
Transcription factors associated with DRGX_PROP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
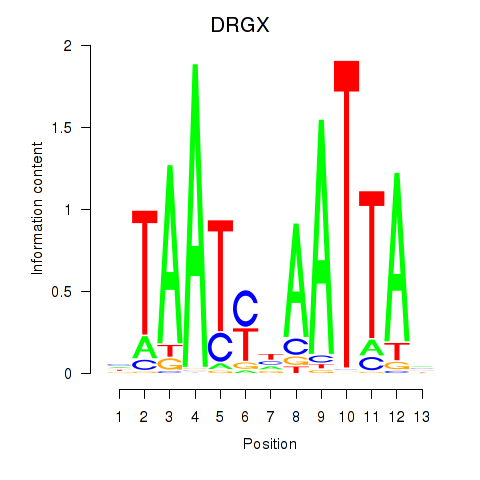
DRGX
|
ENSG00000165606.10 | DRGX |
|
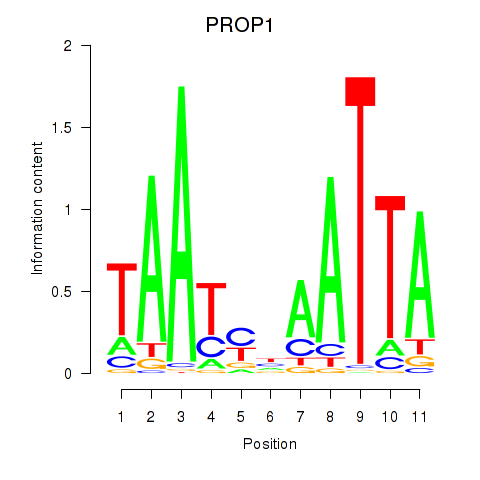
PROP1
|
ENSG00000175325.2 | PROP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROP1 | hg38_v1_chr5_-_177996242_177996242 | -0.59 | 6.1e-04 | Click! |
Activity profile of DRGX_PROP1 motif
Sorted Z-values of DRGX_PROP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DRGX_PROP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33235987 | 12.16 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr16_-_53052849 | 10.80 |
ENST00000619363.2
|
ENSG00000277639.2
|
novel protein |
| chr13_+_50015254 | 5.70 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr9_-_34397800 | 4.49 |
ENST00000297623.7
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr4_-_99435396 | 4.48 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_99435336 | 4.29 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_99435134 | 4.17 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_+_94974984 | 3.88 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr5_-_55173173 | 3.15 |
ENST00000296733.5
ENST00000322374.10 ENST00000381375.7 |
CDC20B
|
cell division cycle 20B |
| chr14_+_75069577 | 3.12 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr5_-_160852200 | 2.91 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr5_-_138139382 | 2.86 |
ENST00000265191.4
|
NME5
|
NME/NM23 family member 5 |
| chrX_+_35919725 | 2.83 |
ENST00000297866.9
ENST00000378653.8 |
CFAP47
|
cilia and flagella associated protein 47 |
| chr6_+_87407965 | 2.81 |
ENST00000369562.9
|
CFAP206
|
cilia and flagella associated protein 206 |
| chr1_-_91906280 | 2.65 |
ENST00000370399.6
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr9_-_123184233 | 2.30 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr8_-_10655137 | 2.21 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chr5_+_36606355 | 2.20 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr5_+_90640718 | 2.20 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr6_-_75363003 | 2.04 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_-_60141004 | 1.78 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr5_+_141192330 | 1.77 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr6_-_32941018 | 1.70 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr5_-_111976925 | 1.67 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr18_-_55321986 | 1.65 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr18_-_55322215 | 1.64 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr6_-_134318097 | 1.64 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_70075317 | 1.59 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr1_+_183805105 | 1.59 |
ENST00000360851.4
|
RGL1
|
ral guanine nucleotide dissociation stimulator like 1 |
| chr17_+_70075215 | 1.59 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr8_-_26867267 | 1.54 |
ENST00000380573.4
|
ADRA1A
|
adrenoceptor alpha 1A |
| chr10_+_68106109 | 1.52 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr8_+_66126896 | 1.51 |
ENST00000353317.9
|
TRIM55
|
tripartite motif containing 55 |
| chr2_+_161416273 | 1.50 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr2_-_218010202 | 1.46 |
ENST00000646520.1
|
TNS1
|
tensin 1 |
| chr3_-_3109980 | 1.43 |
ENST00000256452.7
ENST00000311981.12 ENST00000430514.6 ENST00000456302.5 |
IL5RA
|
interleukin 5 receptor subunit alpha |
| chr3_+_138621207 | 1.38 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_-_52678560 | 1.37 |
ENST00000562351.2
ENST00000261844.11 ENST00000399202.8 ENST00000562135.5 |
FAM214A
|
family with sequence similarity 214 member A |
| chr7_+_90709530 | 1.33 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr5_+_142770367 | 1.29 |
ENST00000645722.2
ENST00000274498.9 |
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr5_+_69565122 | 1.25 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr3_+_138621225 | 1.24 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_-_26572254 | 1.20 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr12_+_75391078 | 1.17 |
ENST00000550916.6
ENST00000378692.7 ENST00000320460.8 ENST00000547164.1 |
GLIPR1L2
|
GLIPR1 like 2 |
| chr18_+_58196736 | 1.17 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr11_-_96343170 | 1.16 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr6_+_116369837 | 1.15 |
ENST00000645988.1
|
DSE
|
dermatan sulfate epimerase |
| chr3_-_27722316 | 1.13 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr12_-_9999176 | 1.12 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr1_+_244051275 | 1.10 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr17_+_55264952 | 1.09 |
ENST00000226067.10
|
HLF
|
HLF transcription factor, PAR bZIP family member |
| chr12_-_49187369 | 1.08 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr2_+_178320228 | 1.04 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr14_-_94770102 | 1.04 |
ENST00000238558.5
|
GSC
|
goosecoid homeobox |
| chr6_-_111483700 | 0.99 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr11_-_26567087 | 0.98 |
ENST00000436318.6
ENST00000281268.12 |
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26572130 | 0.97 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr17_+_73232601 | 0.93 |
ENST00000359042.6
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr7_-_120858066 | 0.92 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr22_+_25219633 | 0.90 |
ENST00000398215.3
|
CRYBB2
|
crystallin beta B2 |
| chr7_-_120857124 | 0.89 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr11_-_26572102 | 0.86 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr5_-_111758061 | 0.86 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr17_+_73232400 | 0.85 |
ENST00000535032.7
ENST00000577615.5 ENST00000585109.5 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr15_+_43800586 | 0.83 |
ENST00000442995.4
ENST00000458412.2 |
HYPK
|
huntingtin interacting protein K |
| chr17_+_44957907 | 0.80 |
ENST00000678938.1
|
NMT1
|
N-myristoyltransferase 1 |
| chr5_+_98773651 | 0.80 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr7_+_123601836 | 0.79 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr11_+_108116688 | 0.79 |
ENST00000672284.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr7_+_90709816 | 0.79 |
ENST00000436577.3
|
CDK14
|
cyclin dependent kinase 14 |
| chr1_+_14929734 | 0.79 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr7_+_123601815 | 0.78 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr3_-_27722699 | 0.77 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr14_-_75069478 | 0.77 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr2_+_149038685 | 0.75 |
ENST00000409642.8
ENST00000409876.5 |
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr4_+_48483324 | 0.74 |
ENST00000273861.5
|
SLC10A4
|
solute carrier family 10 member 4 |
| chr11_-_57712205 | 0.73 |
ENST00000337672.9
ENST00000431606.4 |
MED19
|
mediator complex subunit 19 |
| chr7_+_90709231 | 0.72 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr7_+_114416286 | 0.67 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2 |
| chr16_+_53099100 | 0.65 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr20_-_35742207 | 0.65 |
ENST00000397370.3
ENST00000528062.7 ENST00000374038.7 ENST00000253363.11 ENST00000361162.10 |
RBM39
|
RNA binding motif protein 39 |
| chr5_+_140827950 | 0.61 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr4_+_94995919 | 0.60 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr17_-_41140487 | 0.60 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr11_+_57712574 | 0.59 |
ENST00000278422.9
ENST00000378312.8 |
TMX2
|
thioredoxin related transmembrane protein 2 |
| chr3_-_191282383 | 0.58 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr12_-_52553139 | 0.56 |
ENST00000267119.6
|
KRT71
|
keratin 71 |
| chr6_+_112236806 | 0.55 |
ENST00000588837.5
ENST00000590293.5 ENST00000585450.5 ENST00000629766.2 ENST00000590804.5 ENST00000590584.4 ENST00000628122.2 ENST00000627025.1 ENST00000590673.5 ENST00000585611.5 ENST00000587816.2 |
LAMA4-AS1
ENSG00000281613.2
|
LAMA4 antisense RNA 1 novel ret finger protein-like 4B |
| chr16_+_68085420 | 0.54 |
ENST00000349223.9
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr19_+_49513353 | 0.54 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr1_+_174964750 | 0.53 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chrX_-_15314543 | 0.52 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr16_+_68085344 | 0.52 |
ENST00000575270.5
ENST00000346183.8 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr3_-_11582330 | 0.52 |
ENST00000451674.6
|
VGLL4
|
vestigial like family member 4 |
| chr17_+_73232637 | 0.51 |
ENST00000268942.12
ENST00000426147.6 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr10_-_101229449 | 0.51 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr3_-_12545499 | 0.51 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr17_+_58166982 | 0.50 |
ENST00000545221.2
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr6_+_3258888 | 0.50 |
ENST00000380305.4
|
PSMG4
|
proteasome assembly chaperone 4 |
| chr1_-_169734064 | 0.50 |
ENST00000333360.12
|
SELE
|
selectin E |
| chr9_+_2159850 | 0.50 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_+_44427591 | 0.49 |
ENST00000558791.5
ENST00000260327.9 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr11_-_40294089 | 0.49 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr19_-_11346228 | 0.48 |
ENST00000588560.5
ENST00000592952.5 |
TMEM205
|
transmembrane protein 205 |
| chr17_+_46511511 | 0.48 |
ENST00000576629.5
|
LRRC37A2
|
leucine rich repeat containing 37 member A2 |
| chr19_-_11346196 | 0.45 |
ENST00000586218.5
|
TMEM205
|
transmembrane protein 205 |
| chrX_+_109535775 | 0.44 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr9_+_2159672 | 0.44 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_154121366 | 0.43 |
ENST00000465093.6
ENST00000496710.5 ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor 26 |
| chr15_+_44427793 | 0.42 |
ENST00000558966.5
ENST00000559793.5 ENST00000558968.1 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr12_-_91179355 | 0.42 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr1_-_165445220 | 0.42 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chrX_-_130268883 | 0.42 |
ENST00000447817.1
ENST00000370978.9 |
ZNF280C
|
zinc finger protein 280C |
| chr1_-_165445088 | 0.40 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr20_+_33562365 | 0.40 |
ENST00000346541.7
ENST00000397800.5 ENST00000492345.5 |
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr15_+_57219411 | 0.40 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr1_+_207325629 | 0.39 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr3_-_197573323 | 0.39 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr20_-_18413216 | 0.39 |
ENST00000480488.2
|
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr3_-_186544377 | 0.39 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S |
| chr3_+_156142962 | 0.38 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr16_+_68085552 | 0.38 |
ENST00000329524.8
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr7_-_25228485 | 0.37 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr3_+_149474688 | 0.37 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr2_-_201739205 | 0.37 |
ENST00000681558.1
ENST00000681495.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
| chr11_+_55262152 | 0.36 |
ENST00000417545.5
|
TRIM48
|
tripartite motif containing 48 |
| chr2_+_108621260 | 0.35 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr9_+_101398841 | 0.33 |
ENST00000339664.7
ENST00000374861.7 ENST00000259395.4 ENST00000615466.1 |
ZNF189
|
zinc finger protein 189 |
| chr1_+_42380772 | 0.33 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr7_-_93148345 | 0.32 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr11_-_124320197 | 0.31 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr14_-_37595224 | 0.31 |
ENST00000250448.5
|
FOXA1
|
forkhead box A1 |
| chrY_-_6874027 | 0.31 |
ENST00000215479.10
|
AMELY
|
amelogenin Y-linked |
| chr4_+_112647059 | 0.30 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr5_-_161546708 | 0.30 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr16_+_24537693 | 0.29 |
ENST00000564314.5
ENST00000567686.5 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr5_-_60488055 | 0.29 |
ENST00000505507.6
ENST00000515835.2 ENST00000502484.6 |
PDE4D
|
phosphodiesterase 4D |
| chr5_+_173918186 | 0.29 |
ENST00000657000.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_+_70028452 | 0.28 |
ENST00000530128.5
ENST00000381057.3 ENST00000673563.1 |
HTN3
|
histatin 3 |
| chr11_-_4288083 | 0.28 |
ENST00000638166.1
|
SSU72P4
|
SSU72 pseudogene 4 |
| chr1_+_248508073 | 0.28 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr4_+_159267737 | 0.27 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr8_-_56211257 | 0.27 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr16_-_69339493 | 0.26 |
ENST00000562595.5
ENST00000615447.1 ENST00000306875.10 ENST00000562081.2 |
COG8
|
component of oligomeric golgi complex 8 |
| chr13_+_53028806 | 0.26 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr13_-_94479671 | 0.26 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr9_+_123033660 | 0.26 |
ENST00000616002.3
|
GPR21
|
G protein-coupled receptor 21 |
| chrX_+_69616067 | 0.26 |
ENST00000338901.4
ENST00000525810.5 ENST00000527388.5 ENST00000374553.6 ENST00000374552.9 ENST00000524573.5 |
EDA
|
ectodysplasin A |
| chr2_+_10368764 | 0.26 |
ENST00000620771.4
|
HPCAL1
|
hippocalcin like 1 |
| chr21_-_14210948 | 0.25 |
ENST00000681601.1
|
LIPI
|
lipase I |
| chr7_-_7535863 | 0.25 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr2_-_58241259 | 0.25 |
ENST00000481670.2
ENST00000403676.5 ENST00000427708.6 ENST00000403295.7 ENST00000233741.9 ENST00000446381.5 ENST00000417361.1 ENST00000402135.7 ENST00000449070.5 |
FANCL
|
FA complementation group L |
| chr3_-_158106408 | 0.25 |
ENST00000483851.7
|
SHOX2
|
short stature homeobox 2 |
| chr3_+_2892199 | 0.24 |
ENST00000397459.6
|
CNTN4
|
contactin 4 |
| chr15_+_48191648 | 0.24 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr5_-_67196791 | 0.23 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr21_-_14210884 | 0.23 |
ENST00000679868.1
ENST00000400211.3 ENST00000680801.1 ENST00000536861.6 ENST00000614229.5 |
LIPI
|
lipase I |
| chr6_-_136550407 | 0.23 |
ENST00000354570.8
|
MAP7
|
microtubule associated protein 7 |
| chr6_-_41071825 | 0.22 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr3_-_146528750 | 0.22 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr2_+_10368645 | 0.21 |
ENST00000613496.4
|
HPCAL1
|
hippocalcin like 1 |
| chr12_-_118359105 | 0.19 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr3_-_9249623 | 0.18 |
ENST00000383836.8
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr11_+_5351508 | 0.17 |
ENST00000380219.1
|
OR51B6
|
olfactory receptor family 51 subfamily B member 6 |
| chr17_-_41149823 | 0.17 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr3_+_111978996 | 0.16 |
ENST00000273359.8
ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10, depalmitoylase |
| chr2_+_36696686 | 0.16 |
ENST00000379242.7
ENST00000389975.7 |
VIT
|
vitrin |
| chr5_-_77638713 | 0.15 |
ENST00000306422.5
|
OTP
|
orthopedia homeobox |
| chr1_+_151254709 | 0.15 |
ENST00000368881.8
ENST00000368884.8 |
PSMD4
|
proteasome 26S subunit, non-ATPase 4 |
| chr3_+_46354072 | 0.15 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chr3_-_18438767 | 0.15 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr1_-_219613069 | 0.15 |
ENST00000367211.6
ENST00000651890.1 |
ZC3H11B
|
zinc finger CCCH-type containing 11B |
| chr11_-_95910665 | 0.14 |
ENST00000674610.1
ENST00000675660.1 |
MTMR2
|
myotubularin related protein 2 |
| chr12_+_19205294 | 0.14 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr13_-_40982880 | 0.14 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr11_-_95910748 | 0.13 |
ENST00000675933.1
|
MTMR2
|
myotubularin related protein 2 |
| chr8_-_81483226 | 0.12 |
ENST00000256104.5
|
FABP4
|
fatty acid binding protein 4 |
| chr14_+_22516273 | 0.12 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr8_-_20183090 | 0.12 |
ENST00000265808.11
ENST00000522513.5 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr5_-_116536458 | 0.12 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A |
| chr16_+_86566821 | 0.12 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr1_-_50423431 | 0.12 |
ENST00000404795.4
|
DMRTA2
|
DMRT like family A2 |
| chr5_-_161546671 | 0.11 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr14_+_20110739 | 0.11 |
ENST00000641386.2
ENST00000641633.2 |
OR4K17
|
olfactory receptor family 4 subfamily K member 17 |
| chr17_-_29005913 | 0.10 |
ENST00000442608.7
ENST00000317338.17 ENST00000335960.10 |
SEZ6
|
seizure related 6 homolog |
| chr11_+_55635113 | 0.10 |
ENST00000641760.1
|
OR4P4
|
olfactory receptor family 4 subfamily P member 4 |
| chr8_-_20183127 | 0.10 |
ENST00000276373.10
ENST00000519026.5 ENST00000440926.3 ENST00000437980.3 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr4_+_164877164 | 0.09 |
ENST00000507152.6
ENST00000515275.1 |
APELA
|
apelin receptor early endogenous ligand |
| chr9_+_65700287 | 0.09 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr7_+_148590760 | 0.09 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chrX_+_101623121 | 0.08 |
ENST00000491568.6
ENST00000479298.5 ENST00000471229.7 |
ARMCX3
|
armadillo repeat containing X-linked 3 |
| chr8_-_17895487 | 0.08 |
ENST00000427924.5
ENST00000381841.4 |
FGL1
|
fibrinogen like 1 |
| chr6_-_69699124 | 0.07 |
ENST00000651675.1
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr8_-_17895403 | 0.07 |
ENST00000381840.5
ENST00000398054.5 |
FGL1
|
fibrinogen like 1 |
| chr11_+_85648018 | 0.07 |
ENST00000532180.1
|
TMEM126A
|
transmembrane protein 126A |
| chr17_-_40755328 | 0.06 |
ENST00000312150.5
|
KRT25
|
keratin 25 |
| chr3_-_157160751 | 0.06 |
ENST00000461804.5
|
CCNL1
|
cyclin L1 |
| chr11_-_102705737 | 0.06 |
ENST00000260229.5
|
MMP27
|
matrix metallopeptidase 27 |
| chr12_+_102957666 | 0.05 |
ENST00000266744.4
|
ASCL1
|
achaete-scute family bHLH transcription factor 1 |
| chr9_-_27005659 | 0.05 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 3.0 | 12.2 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.7 | 2.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.6 | 4.5 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.6 | 1.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.6 | 1.7 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.5 | 1.5 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.5 | 1.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.4 | 1.8 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 6.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 1.0 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.3 | 1.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 0.8 | GO:0072019 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.2 | 0.7 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.2 | 2.2 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.8 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.2 | 1.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.5 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 2.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.3 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 2.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 1.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 5.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.3 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.4 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 2.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 3.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 4.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 1.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0072144 | positive regulation of vascular wound healing(GO:0035470) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 3.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.0 | GO:2000405 | negative regulation of T cell migration(GO:2000405) |
| 0.0 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 1.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 1.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.0 | GO:0071284 | epinephrine metabolic process(GO:0042414) copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 3.2 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.7 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 1.3 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0097545 | radial spoke(GO:0001534) axonemal outer doublet(GO:0097545) |
| 0.4 | 2.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 2.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 4.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 3.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 3.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 3.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0098576 | lumenal side of membrane(GO:0098576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.9 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.5 | 1.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 1.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.4 | 4.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 3.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 1.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 1.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 2.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 0.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.8 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.2 | 0.8 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 3.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 2.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 3.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 2.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 2.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 1.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 2.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.7 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 1.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 3.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 3.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 4.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 12.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 4.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.3 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |