Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
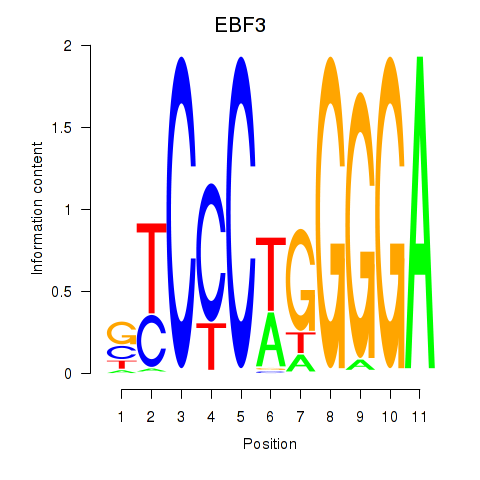
Results for EBF3
Z-value: 0.60
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.16 | EBF3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EBF3 | hg38_v1_chr10_-_129964240_129964291 | -0.17 | 3.7e-01 | Click! |
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_111255982 | 1.17 |
ENST00000637637.1
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr22_-_37244237 | 1.05 |
ENST00000401529.3
ENST00000249071.11 |
RAC2
|
Rac family small GTPase 2 |
| chrX_+_136205982 | 1.01 |
ENST00000628568.1
|
FHL1
|
four and a half LIM domains 1 |
| chr22_-_37244417 | 0.96 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chr5_+_176810552 | 0.90 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A |
| chr16_-_85750951 | 0.89 |
ENST00000602675.5
|
C16orf74
|
chromosome 16 open reading frame 74 |
| chrX_+_70290077 | 0.88 |
ENST00000374403.4
|
KIF4A
|
kinesin family member 4A |
| chr5_+_176810498 | 0.88 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A |
| chr16_-_85751112 | 0.86 |
ENST00000602766.1
|
C16orf74
|
chromosome 16 open reading frame 74 |
| chr15_+_90184912 | 0.79 |
ENST00000561085.1
ENST00000332496.10 |
SEMA4B
|
semaphorin 4B |
| chr21_-_44261854 | 0.77 |
ENST00000431166.1
|
DNMT3L
|
DNA methyltransferase 3 like |
| chr5_-_147453888 | 0.77 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr16_-_85751028 | 0.76 |
ENST00000284245.9
ENST00000602914.1 |
C16orf74
|
chromosome 16 open reading frame 74 |
| chrX_+_136147465 | 0.73 |
ENST00000651929.2
|
FHL1
|
four and a half LIM domains 1 |
| chr17_+_41819201 | 0.67 |
ENST00000455106.1
|
FKBP10
|
FKBP prolyl isomerase 10 |
| chrX_+_136147525 | 0.67 |
ENST00000652745.1
ENST00000627578.2 ENST00000652457.1 ENST00000394155.8 ENST00000618438.4 |
FHL1
|
four and a half LIM domains 1 |
| chr17_-_41397600 | 0.64 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr1_-_41484680 | 0.63 |
ENST00000372587.5
|
EDN2
|
endothelin 2 |
| chr3_-_59049947 | 0.62 |
ENST00000491845.5
ENST00000472469.5 ENST00000482387.6 ENST00000295966.11 |
CFAP20DC
|
CFAP20 domain containing |
| chr5_+_155013755 | 0.62 |
ENST00000435029.6
|
KIF4B
|
kinesin family member 4B |
| chr19_-_18940289 | 0.61 |
ENST00000542541.6
ENST00000433218.6 |
HOMER3
|
homer scaffold protein 3 |
| chrX_+_136147556 | 0.58 |
ENST00000651089.1
ENST00000420362.5 |
FHL1
|
four and a half LIM domains 1 |
| chr14_+_24398986 | 0.55 |
ENST00000382554.4
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr3_+_100492548 | 0.55 |
ENST00000323523.8
ENST00000403410.5 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr12_+_119178920 | 0.55 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr7_+_48088596 | 0.54 |
ENST00000416681.5
ENST00000331803.8 ENST00000432131.5 |
UPP1
|
uridine phosphorylase 1 |
| chr14_-_94129577 | 0.54 |
ENST00000238609.4
|
IFI27L2
|
interferon alpha inducible protein 27 like 2 |
| chr18_+_36544544 | 0.52 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr2_+_219460719 | 0.51 |
ENST00000396688.5
|
SPEG
|
striated muscle enriched protein kinase |
| chr14_-_106335613 | 0.51 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr13_+_31846713 | 0.51 |
ENST00000645780.1
|
FRY
|
FRY microtubule binding protein |
| chr9_-_94328644 | 0.51 |
ENST00000341207.5
ENST00000253262.9 |
NUTM2F
|
NUT family member 2F |
| chr19_+_41708635 | 0.50 |
ENST00000617332.4
ENST00000615021.4 ENST00000616453.1 ENST00000405816.5 ENST00000435837.2 |
CEACAM5
ENSG00000267881.2
|
CEA cell adhesion molecule 5 novel protein, readthrough between CEACAM5-CEACAM6 |
| chr1_-_159954213 | 0.50 |
ENST00000368092.7
ENST00000368093.4 |
SLAMF9
|
SLAM family member 9 |
| chr14_+_54567612 | 0.50 |
ENST00000251091.9
ENST00000392067.7 ENST00000631086.2 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr9_+_125747345 | 0.49 |
ENST00000342287.9
ENST00000373489.10 ENST00000373487.8 |
PBX3
|
PBX homeobox 3 |
| chr17_+_2056073 | 0.48 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chr10_-_13972355 | 0.48 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr9_+_125748175 | 0.45 |
ENST00000491787.7
ENST00000447726.6 |
PBX3
|
PBX homeobox 3 |
| chr11_-_107858777 | 0.44 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr2_-_89143133 | 0.43 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr16_+_67199104 | 0.42 |
ENST00000360833.6
ENST00000652269.1 ENST00000393997.8 |
ELMO3
|
engulfment and cell motility 3 |
| chr3_-_195811889 | 0.42 |
ENST00000475231.5
|
MUC4
|
mucin 4, cell surface associated |
| chr19_+_46602050 | 0.41 |
ENST00000599839.5
ENST00000596362.1 |
CALM3
|
calmodulin 3 |
| chr2_+_219461255 | 0.41 |
ENST00000396686.5
ENST00000396689.2 |
SPEG
|
striated muscle enriched protein kinase |
| chr7_+_77538027 | 0.40 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr14_-_106360320 | 0.39 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chrX_+_38561530 | 0.39 |
ENST00000378482.7
ENST00000286824.6 |
TSPAN7
|
tetraspanin 7 |
| chr12_+_119178953 | 0.39 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr16_-_90008988 | 0.38 |
ENST00000568662.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr14_-_106130061 | 0.38 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr1_-_159945596 | 0.38 |
ENST00000361509.7
ENST00000611023.1 ENST00000368094.6 |
IGSF9
|
immunoglobulin superfamily member 9 |
| chr6_-_30687200 | 0.37 |
ENST00000399199.7
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr10_+_100347225 | 0.37 |
ENST00000370355.3
|
SCD
|
stearoyl-CoA desaturase |
| chr21_+_36135071 | 0.37 |
ENST00000290354.6
|
CBR3
|
carbonyl reductase 3 |
| chr11_+_124865425 | 0.37 |
ENST00000397801.6
|
ROBO3
|
roundabout guidance receptor 3 |
| chr19_-_14114156 | 0.37 |
ENST00000589994.6
|
PRKACA
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr17_-_7590072 | 0.37 |
ENST00000538513.6
ENST00000570788.1 ENST00000250055.3 |
SOX15
|
SRY-box transcription factor 15 |
| chr6_-_10419638 | 0.37 |
ENST00000319516.8
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr10_+_97572771 | 0.36 |
ENST00000370655.6
ENST00000455090.1 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr3_+_50269140 | 0.36 |
ENST00000616701.5
ENST00000433753.4 ENST00000611067.4 |
SEMA3B
|
semaphorin 3B |
| chr7_+_77538059 | 0.36 |
ENST00000435495.6
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr5_+_150190035 | 0.36 |
ENST00000230671.7
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 member 7 |
| chr10_+_97572493 | 0.35 |
ENST00000307518.9
ENST00000298808.9 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr2_+_31234144 | 0.35 |
ENST00000322054.10
|
EHD3
|
EH domain containing 3 |
| chr3_+_52794768 | 0.34 |
ENST00000621946.4
ENST00000416872.6 ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr3_-_195811916 | 0.34 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated |
| chr20_+_43667019 | 0.34 |
ENST00000396863.8
|
MYBL2
|
MYB proto-oncogene like 2 |
| chr2_+_89947508 | 0.34 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr13_-_30307539 | 0.33 |
ENST00000380615.8
|
KATNAL1
|
katanin catalytic subunit A1 like 1 |
| chr3_-_195811857 | 0.32 |
ENST00000349607.8
ENST00000346145.8 |
MUC4
|
mucin 4, cell surface associated |
| chr1_-_6419903 | 0.32 |
ENST00000377836.8
ENST00000487437.5 ENST00000489730.1 ENST00000377834.8 |
HES2
|
hes family bHLH transcription factor 2 |
| chr12_-_109833373 | 0.32 |
ENST00000261740.7
|
TRPV4
|
transient receptor potential cation channel subfamily V member 4 |
| chr16_+_71358713 | 0.32 |
ENST00000349553.9
ENST00000302628.9 ENST00000562305.5 |
CALB2
|
calbindin 2 |
| chr5_+_141402764 | 0.32 |
ENST00000573521.2
ENST00000616887.1 |
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr20_+_43667105 | 0.32 |
ENST00000217026.5
|
MYBL2
|
MYB proto-oncogene like 2 |
| chr11_+_64291285 | 0.31 |
ENST00000422670.7
ENST00000538767.1 |
KCNK4
|
potassium two pore domain channel subfamily K member 4 |
| chr10_+_110644306 | 0.31 |
ENST00000369519.4
|
RBM20
|
RNA binding motif protein 20 |
| chr19_-_2042066 | 0.31 |
ENST00000591588.1
ENST00000591142.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr7_-_101237791 | 0.30 |
ENST00000308344.10
|
CLDN15
|
claudin 15 |
| chr11_-_130428605 | 0.30 |
ENST00000257359.7
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif 8 |
| chr11_+_118956289 | 0.30 |
ENST00000264031.3
|
UPK2
|
uroplakin 2 |
| chr14_+_78403686 | 0.30 |
ENST00000553631.1
ENST00000554719.5 |
NRXN3
|
neurexin 3 |
| chr12_+_53050014 | 0.30 |
ENST00000314250.11
|
TNS2
|
tensin 2 |
| chr19_+_41708585 | 0.29 |
ENST00000398599.8
ENST00000221992.11 |
CEACAM5
|
CEA cell adhesion molecule 5 |
| chr3_-_142888896 | 0.29 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr14_-_67695747 | 0.29 |
ENST00000553384.5
ENST00000381346.9 ENST00000557726.1 |
RDH11
|
retinol dehydrogenase 11 |
| chr5_+_149271848 | 0.29 |
ENST00000296721.9
|
AFAP1L1
|
actin filament associated protein 1 like 1 |
| chr3_-_52239082 | 0.29 |
ENST00000499914.2
ENST00000678838.1 ENST00000305533.10 ENST00000678330.1 |
TWF2
|
twinfilin actin binding protein 2 |
| chr9_+_96928843 | 0.29 |
ENST00000372322.4
|
NUTM2G
|
NUT family member 2G |
| chr1_+_15684284 | 0.29 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chrX_+_48508949 | 0.28 |
ENST00000359882.8
ENST00000326194.11 ENST00000355961.8 ENST00000683923.1 ENST00000489940.5 ENST00000361988.7 |
PORCN
|
porcupine O-acyltransferase |
| chr10_-_100330220 | 0.28 |
ENST00000318222.4
|
PKD2L1
|
polycystin 2 like 1, transient receptor potential cation channel |
| chr9_+_132978687 | 0.28 |
ENST00000372122.4
ENST00000372123.5 |
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr12_+_7130341 | 0.27 |
ENST00000266546.11
|
CLSTN3
|
calsyntenin 3 |
| chr9_+_132978651 | 0.27 |
ENST00000636137.1
|
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr12_+_53050179 | 0.27 |
ENST00000546602.5
ENST00000552570.5 ENST00000549700.5 |
TNS2
|
tensin 2 |
| chr2_-_89027700 | 0.27 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr7_-_44141074 | 0.27 |
ENST00000457314.5
ENST00000447951.1 ENST00000431007.1 |
MYL7
|
myosin light chain 7 |
| chr22_+_23980123 | 0.27 |
ENST00000621118.4
|
GSTT2
|
glutathione S-transferase theta 2 (gene/pseudogene) |
| chr1_-_154970735 | 0.27 |
ENST00000368445.9
ENST00000448116.7 ENST00000368449.8 |
SHC1
|
SHC adaptor protein 1 |
| chr7_-_100642721 | 0.26 |
ENST00000462107.1
|
TFR2
|
transferrin receptor 2 |
| chr3_-_186108501 | 0.26 |
ENST00000422039.1
ENST00000434744.5 |
ETV5
|
ETS variant transcription factor 5 |
| chr1_-_156859087 | 0.26 |
ENST00000368195.4
|
INSRR
|
insulin receptor related receptor |
| chr17_+_40309161 | 0.26 |
ENST00000254066.10
|
RARA
|
retinoic acid receptor alpha |
| chr16_+_67199509 | 0.26 |
ENST00000477898.5
|
ELMO3
|
engulfment and cell motility 3 |
| chr2_-_75569711 | 0.26 |
ENST00000233712.5
|
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chr17_-_41918966 | 0.25 |
ENST00000537919.5
ENST00000353196.5 ENST00000393896.6 |
ACLY
|
ATP citrate lyase |
| chr2_-_218286763 | 0.25 |
ENST00000444881.5
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr17_-_41918944 | 0.25 |
ENST00000352035.7
ENST00000590770.5 ENST00000590151.5 |
ACLY
|
ATP citrate lyase |
| chr5_-_172771187 | 0.25 |
ENST00000239223.4
|
DUSP1
|
dual specificity phosphatase 1 |
| chr12_-_122730828 | 0.25 |
ENST00000432564.3
|
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr19_-_38253238 | 0.25 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr3_-_142889156 | 0.24 |
ENST00000648195.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr17_-_76027212 | 0.24 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr7_-_100100716 | 0.24 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr15_+_63042632 | 0.24 |
ENST00000288398.10
ENST00000358278.7 ENST00000610733.1 ENST00000403994.9 ENST00000357980.9 ENST00000559556.5 ENST00000267996.11 ENST00000559397.6 ENST00000561266.6 ENST00000560970.6 |
TPM1
|
tropomyosin 1 |
| chr1_+_1033987 | 0.24 |
ENST00000651234.1
ENST00000652369.1 |
AGRN
|
agrin |
| chr3_-_142889075 | 0.24 |
ENST00000295992.8
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr15_-_38564635 | 0.24 |
ENST00000450598.6
ENST00000559830.5 ENST00000558164.5 ENST00000539159.5 ENST00000310803.10 |
RASGRP1
|
RAS guanyl releasing protein 1 |
| chrX_-_154409278 | 0.23 |
ENST00000369808.7
|
DNASE1L1
|
deoxyribonuclease 1 like 1 |
| chr7_-_44141285 | 0.23 |
ENST00000458240.5
ENST00000223364.7 |
MYL7
|
myosin light chain 7 |
| chr10_+_97584347 | 0.23 |
ENST00000370649.3
ENST00000370646.9 |
ENSG00000249967.1
HOGA1
|
novel protein 4-hydroxy-2-oxoglutarate aldolase 1 |
| chr9_+_130200375 | 0.23 |
ENST00000630865.1
|
NCS1
|
neuronal calcium sensor 1 |
| chr17_-_76027296 | 0.23 |
ENST00000301607.8
|
EVPL
|
envoplakin |
| chr19_-_40257045 | 0.22 |
ENST00000578615.6
|
AKT2
|
AKT serine/threonine kinase 2 |
| chr3_-_46882106 | 0.22 |
ENST00000662933.1
|
MYL3
|
myosin light chain 3 |
| chr11_+_64234569 | 0.22 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr16_-_67833842 | 0.22 |
ENST00000566758.5
ENST00000626059.2 ENST00000564817.5 |
CENPT
|
centromere protein T |
| chrX_-_154409246 | 0.22 |
ENST00000369807.6
|
DNASE1L1
|
deoxyribonuclease 1 like 1 |
| chr8_+_84184875 | 0.22 |
ENST00000517638.5
ENST00000522647.1 |
RALYL
|
RALY RNA binding protein like |
| chr9_+_113594118 | 0.22 |
ENST00000620489.1
|
RGS3
|
regulator of G protein signaling 3 |
| chr12_-_48865863 | 0.22 |
ENST00000309739.6
|
RND1
|
Rho family GTPase 1 |
| chr17_-_81923532 | 0.21 |
ENST00000392366.7
|
MAFG
|
MAF bZIP transcription factor G |
| chr3_-_190122317 | 0.21 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr3_+_52779916 | 0.21 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr19_-_10517439 | 0.21 |
ENST00000333430.6
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr1_+_161749762 | 0.21 |
ENST00000367943.5
|
DUSP12
|
dual specificity phosphatase 12 |
| chr3_+_46882180 | 0.21 |
ENST00000427125.6
ENST00000430002.6 |
PTH1R
|
parathyroid hormone 1 receptor |
| chr11_+_75562274 | 0.21 |
ENST00000532356.5
ENST00000524558.5 |
SERPINH1
|
serpin family H member 1 |
| chr17_+_7445055 | 0.21 |
ENST00000306071.7
ENST00000572857.5 |
CHRNB1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr12_-_663572 | 0.21 |
ENST00000662884.1
|
NINJ2
|
ninjurin 2 |
| chr2_+_87338511 | 0.21 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr3_-_37176305 | 0.21 |
ENST00000440230.5
ENST00000421276.6 ENST00000354379.8 |
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr11_+_75562242 | 0.20 |
ENST00000526397.5
ENST00000529643.1 ENST00000525492.5 ENST00000530284.5 ENST00000358171.8 |
SERPINH1
|
serpin family H member 1 |
| chr19_+_41443130 | 0.20 |
ENST00000378187.3
|
ERICH4
|
glutamate rich 4 |
| chr1_-_116667668 | 0.20 |
ENST00000369486.8
ENST00000369483.5 |
IGSF3
|
immunoglobulin superfamily member 3 |
| chr11_-_126211637 | 0.20 |
ENST00000533628.5
ENST00000298317.9 ENST00000532674.2 |
RPUSD4
|
RNA pseudouridine synthase D4 |
| chr22_+_20967243 | 0.20 |
ENST00000683034.1
ENST00000440238.4 ENST00000405089.5 |
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr1_+_243256034 | 0.20 |
ENST00000366541.8
|
SDCCAG8
|
SHH signaling and ciliogenesis regulator SDCCAG8 |
| chr22_+_20967212 | 0.20 |
ENST00000434714.6
|
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr3_-_128467248 | 0.20 |
ENST00000319153.3
|
DNAJB8
|
DnaJ heat shock protein family (Hsp40) member B8 |
| chr1_+_247416166 | 0.20 |
ENST00000391827.3
ENST00000336119.8 |
NLRP3
|
NLR family pyrin domain containing 3 |
| chr22_-_37427433 | 0.20 |
ENST00000452946.1
ENST00000402918.7 |
ELFN2
ELFN2
|
extracellular leucine rich repeat and fibronectin type III domain containing 2 extracellular leucine rich repeat and fibronectin type III domain containing 2 |
| chr6_+_31587268 | 0.20 |
ENST00000396101.7
ENST00000490742.5 |
LST1
|
leukocyte specific transcript 1 |
| chr16_+_30949054 | 0.20 |
ENST00000318663.5
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr12_-_663431 | 0.20 |
ENST00000305108.10
|
NINJ2
|
ninjurin 2 |
| chr1_+_247416149 | 0.19 |
ENST00000366497.6
ENST00000391828.8 |
NLRP3
|
NLR family pyrin domain containing 3 |
| chr5_+_149271906 | 0.19 |
ENST00000515000.1
|
AFAP1L1
|
actin filament associated protein 1 like 1 |
| chr19_-_7702139 | 0.19 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II |
| chrX_-_52654900 | 0.19 |
ENST00000298181.6
|
SSX7
|
SSX family member 7 |
| chr3_+_194136138 | 0.19 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr17_-_1492660 | 0.19 |
ENST00000648651.1
|
MYO1C
|
myosin IC |
| chr11_+_75562056 | 0.19 |
ENST00000533603.5
|
SERPINH1
|
serpin family H member 1 |
| chr1_-_24112125 | 0.19 |
ENST00000374434.4
|
MYOM3
|
myomesin 3 |
| chr18_+_31498168 | 0.19 |
ENST00000261590.13
ENST00000585206.1 ENST00000683654.1 |
DSG2
|
desmoglein 2 |
| chr19_-_7702124 | 0.18 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr20_+_63865228 | 0.18 |
ENST00000611972.4
ENST00000615907.4 ENST00000369927.8 ENST00000346249.9 ENST00000348257.9 ENST00000352482.8 ENST00000351424.8 ENST00000217121.9 ENST00000358548.4 |
TPD52L2
|
TPD52 like 2 |
| chr17_-_81835042 | 0.18 |
ENST00000330261.5
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1 regulatory subunit 27 |
| chr11_+_61228377 | 0.18 |
ENST00000537932.5
|
PGA4
|
pepsinogen A4 |
| chr16_+_25691953 | 0.18 |
ENST00000331351.6
|
HS3ST4
|
heparan sulfate-glucosamine 3-sulfotransferase 4 |
| chr12_+_8509460 | 0.18 |
ENST00000382064.6
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chr11_-_62679055 | 0.18 |
ENST00000294119.6
ENST00000529640.5 ENST00000301935.10 ENST00000534176.1 ENST00000616865.4 |
UBXN1
|
UBX domain protein 1 |
| chr2_+_216412743 | 0.18 |
ENST00000358207.9
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a like 1 |
| chr19_+_33374312 | 0.18 |
ENST00000585933.2
|
CEBPG
|
CCAAT enhancer binding protein gamma |
| chr14_+_18967434 | 0.18 |
ENST00000547889.6
|
POTEM
|
POTE ankyrin domain family member M |
| chr18_+_11981488 | 0.18 |
ENST00000269159.8
|
IMPA2
|
inositol monophosphatase 2 |
| chr3_+_6861107 | 0.18 |
ENST00000357716.9
ENST00000486284.5 ENST00000389336.8 |
GRM7
|
glutamate metabotropic receptor 7 |
| chr4_-_176002332 | 0.18 |
ENST00000280187.11
ENST00000512509.5 |
GPM6A
|
glycoprotein M6A |
| chr15_+_81000913 | 0.18 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chr8_-_19602484 | 0.18 |
ENST00000454498.6
ENST00000520003.5 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr1_+_12230006 | 0.18 |
ENST00000620676.6
|
VPS13D
|
vacuolar protein sorting 13 homolog D |
| chr16_-_30091226 | 0.18 |
ENST00000279386.6
ENST00000627355.2 |
TBX6
|
T-box transcription factor 6 |
| chr1_-_23368301 | 0.17 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr2_-_128028114 | 0.17 |
ENST00000259234.10
|
SAP130
|
Sin3A associated protein 130 |
| chr17_-_7179544 | 0.17 |
ENST00000619926.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr22_-_28306645 | 0.17 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr2_+_176099787 | 0.17 |
ENST00000406506.4
ENST00000404162.2 |
HOXD12
|
homeobox D12 |
| chr8_+_12945667 | 0.17 |
ENST00000524591.7
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr15_+_75347610 | 0.17 |
ENST00000564784.5
ENST00000569035.5 |
NEIL1
|
nei like DNA glycosylase 1 |
| chr17_+_81098118 | 0.17 |
ENST00000416299.6
|
BAIAP2
|
BAR/IMD domain containing adaptor protein 2 |
| chr19_+_45507470 | 0.17 |
ENST00000245932.11
ENST00000592139.1 ENST00000590603.1 |
VASP
|
vasodilator stimulated phosphoprotein |
| chr1_-_42817357 | 0.17 |
ENST00000372521.9
|
SVBP
|
small vasohibin binding protein |
| chr11_-_67353503 | 0.17 |
ENST00000539074.1
ENST00000530584.5 ENST00000531239.2 ENST00000312419.8 ENST00000529704.5 |
POLD4
|
DNA polymerase delta 4, accessory subunit |
| chrX_+_106693838 | 0.17 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr7_-_101237827 | 0.17 |
ENST00000611078.4
|
CLDN15
|
claudin 15 |
| chr2_-_187513641 | 0.17 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr14_-_100548098 | 0.16 |
ENST00000554356.6
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr16_+_69950705 | 0.16 |
ENST00000615430.4
|
CLEC18A
|
C-type lectin domain family 18 member A |
| chr5_+_181147586 | 0.16 |
ENST00000641492.1
ENST00000641791.1 |
OR2V2
|
olfactory receptor family 2 subfamily V member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.2 | 2.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.2 | 1.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 0.2 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.2 | 0.6 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.2 | 0.5 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.2 | 0.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 0.3 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 0.3 | GO:2000506 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) negative regulation of energy homeostasis(GO:2000506) |
| 0.1 | 0.4 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 1.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.4 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.2 | GO:1990009 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.1 | 0.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.3 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.2 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.8 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.2 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.5 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 1.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 1.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.3 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.0 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.3 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 3.0 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 1.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.5 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 2.8 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071753 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.3 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.5 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 1.9 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0035631 | XY body(GO:0001741) CD40 receptor complex(GO:0035631) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.2 | 0.5 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 1.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.8 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.4 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 1.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0047238 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 2.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |