Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
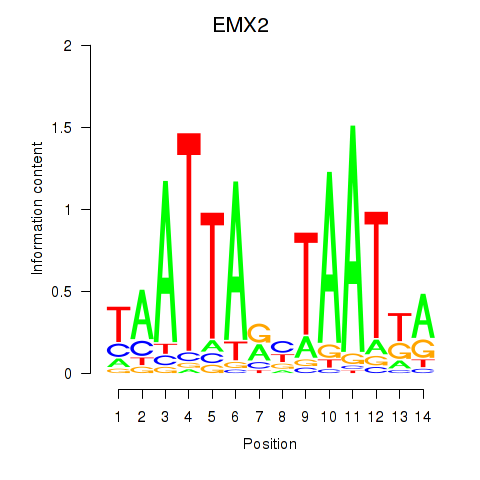
Results for EMX2
Z-value: 1.21
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.12 | EMX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg38_v1_chr10_+_117542416_117542445, hg38_v1_chr10_+_117543567_117543658, hg38_v1_chr10_+_117542721_117542754 | -0.36 | 4.9e-02 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_63652663 | 12.37 |
ENST00000343837.8
ENST00000469440.5 |
SNTN
|
sentan, cilia apical structure protein |
| chr11_-_5227063 | 6.85 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr6_+_32439866 | 6.33 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr2_+_227871618 | 5.74 |
ENST00000309931.3
ENST00000440997.1 |
DAW1
|
dynein assembly factor with WD repeats 1 |
| chr11_-_63608542 | 5.55 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr17_-_76141240 | 5.54 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr7_+_48035511 | 5.28 |
ENST00000420324.5
ENST00000539619.5 ENST00000435376.5 ENST00000430738.5 ENST00000348904.4 |
C7orf57
|
chromosome 7 open reading frame 57 |
| chr5_+_140875299 | 4.92 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chr4_-_88284747 | 4.73 |
ENST00000514204.1
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr21_-_42315336 | 4.71 |
ENST00000398431.2
ENST00000518498.3 |
TFF3
|
trefoil factor 3 |
| chr11_+_27055215 | 4.49 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr5_-_138139382 | 4.49 |
ENST00000265191.4
|
NME5
|
NME/NM23 family member 5 |
| chr5_+_36608146 | 4.13 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr4_-_99435396 | 4.11 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr16_-_28623560 | 4.08 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr9_-_110208156 | 4.02 |
ENST00000400613.5
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr6_-_52840843 | 3.81 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr3_-_112829367 | 3.79 |
ENST00000448932.4
ENST00000617549.3 |
CD200R1L
|
CD200 receptor 1 like |
| chr12_-_68302872 | 3.69 |
ENST00000539972.5
|
MDM1
|
Mdm1 nuclear protein |
| chr4_-_99435336 | 3.62 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_-_26572254 | 3.54 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26572130 | 3.22 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_-_99435134 | 3.05 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_-_26572102 | 3.03 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_119196769 | 3.03 |
ENST00000415318.2
|
CCDC153
|
coiled-coil domain containing 153 |
| chr18_+_63702958 | 3.00 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr8_-_132675533 | 2.92 |
ENST00000620350.5
ENST00000518642.5 ENST00000250173.5 |
LRRC6
|
leucine rich repeat containing 6 |
| chr3_+_72888031 | 2.88 |
ENST00000389617.9
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr6_+_150368892 | 2.80 |
ENST00000229447.9
ENST00000392256.6 |
IYD
|
iodotyrosine deiodinase |
| chr1_+_36084079 | 2.78 |
ENST00000207457.8
|
TEKT2
|
tektin 2 |
| chr8_-_132675567 | 2.70 |
ENST00000519595.5
|
LRRC6
|
leucine rich repeat containing 6 |
| chr3_+_113897470 | 2.68 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr1_+_78649818 | 2.53 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr1_+_170935526 | 2.52 |
ENST00000367758.7
ENST00000367759.9 |
MROH9
|
maestro heat like repeat family member 9 |
| chr12_-_10130241 | 2.46 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr13_-_19503277 | 2.44 |
ENST00000382978.5
ENST00000400230.6 ENST00000255310.10 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr19_+_48695952 | 2.37 |
ENST00000522966.2
ENST00000425340.3 ENST00000391876.5 |
FUT2
|
fucosyltransferase 2 |
| chr3_+_131026844 | 2.28 |
ENST00000510769.5
ENST00000383366.9 ENST00000510688.5 ENST00000511262.5 |
NEK11
|
NIMA related kinase 11 |
| chr5_-_16508788 | 2.27 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr7_+_90709530 | 2.25 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr12_-_10130143 | 2.18 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr1_-_60073750 | 2.14 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr5_+_90640718 | 2.13 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr16_-_28623330 | 2.12 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein |
| chr4_+_68815991 | 2.09 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr4_-_25863537 | 1.98 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr21_-_7825797 | 1.91 |
ENST00000617668.2
|
KCNE1B
|
potassium voltage-gated channel subfamily E regulatory subunit 1B |
| chr7_+_90709231 | 1.88 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr1_-_86383078 | 1.87 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like |
| chr6_-_52763473 | 1.87 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr19_+_49513353 | 1.84 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr5_-_41213505 | 1.82 |
ENST00000337836.10
ENST00000433294.1 |
C6
|
complement C6 |
| chr11_+_66509079 | 1.80 |
ENST00000419755.3
|
ENSG00000256349.1
|
novel protein |
| chr4_+_94974984 | 1.76 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr12_-_10130082 | 1.74 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr14_-_106470788 | 1.74 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr2_-_98663464 | 1.71 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr3_-_122793772 | 1.70 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr5_-_111976925 | 1.64 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr5_+_42548043 | 1.63 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr4_+_41538143 | 1.56 |
ENST00000503057.6
ENST00000511496.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr18_-_55423757 | 1.55 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chr16_+_69565958 | 1.52 |
ENST00000349945.7
ENST00000354436.6 |
NFAT5
|
nuclear factor of activated T cells 5 |
| chr16_-_46748337 | 1.42 |
ENST00000394809.9
|
MYLK3
|
myosin light chain kinase 3 |
| chr2_-_206159194 | 1.33 |
ENST00000455934.6
ENST00000449699.5 ENST00000454195.1 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr6_-_75363003 | 1.30 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_-_109801978 | 1.29 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr12_+_74537787 | 1.28 |
ENST00000519948.4
|
ATXN7L3B
|
ataxin 7 like 3B |
| chr9_-_76906041 | 1.23 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr1_-_34985288 | 1.21 |
ENST00000417456.1
ENST00000373337.3 |
ENSG00000284773.1
TMEM35B
|
novel protein transmembrane protein 35B |
| chr2_-_206159410 | 1.12 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr13_-_70108441 | 1.12 |
ENST00000377844.9
ENST00000545028.2 |
KLHL1
|
kelch like family member 1 |
| chr4_+_73836667 | 1.09 |
ENST00000226317.10
|
CXCL6
|
C-X-C motif chemokine ligand 6 |
| chr6_+_116100813 | 1.09 |
ENST00000419791.3
ENST00000319550.9 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr3_-_142000353 | 1.09 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr1_+_244835616 | 1.08 |
ENST00000366528.3
ENST00000411948.7 |
COX20
|
cytochrome c oxidase assembly factor COX20 |
| chr16_+_31873772 | 1.08 |
ENST00000394846.7
ENST00000300870.15 |
ZNF267
|
zinc finger protein 267 |
| chr17_+_1771688 | 1.07 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr2_+_109794296 | 1.07 |
ENST00000430736.5
ENST00000016946.8 ENST00000441344.1 |
RGPD5
|
RANBP2 like and GRIP domain containing 5 |
| chrX_+_55717796 | 0.98 |
ENST00000262850.7
|
RRAGB
|
Ras related GTP binding B |
| chr7_-_44848021 | 0.97 |
ENST00000349299.7
ENST00000521529.5 ENST00000350771.7 ENST00000222690.10 ENST00000381124.9 ENST00000437072.5 ENST00000446531.1 ENST00000308153.9 |
H2AZ2
|
H2A.Z variant histone 2 |
| chr8_+_109540075 | 0.96 |
ENST00000614147.1
ENST00000337573.10 |
EBAG9
|
estrogen receptor binding site associated antigen 9 |
| chr4_+_55346213 | 0.95 |
ENST00000679836.1
ENST00000264228.9 ENST00000679707.1 |
SRD5A3
ENSG00000288695.1
|
steroid 5 alpha-reductase 3 novel protein, SRD5A3-RP11-177J6.1 readthrough |
| chr19_-_34773184 | 0.94 |
ENST00000588760.1
ENST00000329285.13 ENST00000587354.6 |
ZNF599
|
zinc finger protein 599 |
| chr2_-_162318129 | 0.93 |
ENST00000679938.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr8_+_109540602 | 0.90 |
ENST00000530629.5
ENST00000620557.4 |
EBAG9
|
estrogen receptor binding site associated antigen 9 |
| chr22_+_31754862 | 0.89 |
ENST00000382111.6
ENST00000645407.1 ENST00000646701.1 |
DEPDC5
ENSG00000285404.1
|
DEP domain containing 5, GATOR1 subcomplex subunit novel protein, DEPDC5-YWHAH readthrough |
| chr12_-_100262356 | 0.89 |
ENST00000548313.5
|
DEPDC4
|
DEP domain containing 4 |
| chr3_-_186570308 | 0.88 |
ENST00000446782.5
|
TBCCD1
|
TBCC domain containing 1 |
| chr3_-_49429304 | 0.88 |
ENST00000636166.1
ENST00000273598.8 ENST00000436744.2 |
ENSG00000283189.2
NICN1
|
novel protein nicolin 1 |
| chr1_+_174875505 | 0.86 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr14_+_101809795 | 0.85 |
ENST00000350249.7
ENST00000557621.5 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chr15_-_75455767 | 0.84 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chrX_+_55717733 | 0.84 |
ENST00000414239.5
ENST00000374941.9 |
RRAGB
|
Ras related GTP binding B |
| chr3_-_49429252 | 0.82 |
ENST00000615713.4
|
NICN1
|
nicolin 1 |
| chr20_-_31390580 | 0.82 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chrX_+_106168297 | 0.81 |
ENST00000337685.6
ENST00000357175.6 |
PWWP3B
|
PWWP domain containing 3B |
| chr14_+_21990357 | 0.81 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr8_-_142777802 | 0.80 |
ENST00000621401.4
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr1_-_152115443 | 0.79 |
ENST00000614923.1
|
TCHH
|
trichohyalin |
| chr3_+_132597260 | 0.79 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr20_+_15196834 | 0.79 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr5_-_180861248 | 0.77 |
ENST00000502412.2
ENST00000512132.5 ENST00000506439.5 |
ZFP62
|
ZFP62 zinc finger protein |
| chr1_-_92486916 | 0.76 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr3_-_131026726 | 0.76 |
ENST00000514044.5
ENST00000264992.8 |
ASTE1
|
asteroid homolog 1 |
| chr11_-_129024157 | 0.74 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr12_-_57826295 | 0.73 |
ENST00000549039.5
|
CTDSP2
|
CTD small phosphatase 2 |
| chr10_-_20897288 | 0.71 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr12_-_91180365 | 0.71 |
ENST00000547937.5
|
DCN
|
decorin |
| chr11_+_10455292 | 0.70 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr6_+_28124596 | 0.70 |
ENST00000340487.5
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr2_-_110577101 | 0.69 |
ENST00000330331.9
ENST00000446930.1 ENST00000329516.8 |
RGPD6
|
RANBP2 like and GRIP domain containing 6 |
| chr1_+_207325629 | 0.68 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr16_-_28363508 | 0.68 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family member B6 |
| chr15_-_42208153 | 0.68 |
ENST00000348544.4
ENST00000318006.10 |
VPS39
|
VPS39 subunit of HOPS complex |
| chr9_-_96778053 | 0.67 |
ENST00000375231.5
ENST00000223428.9 |
ZNF510
|
zinc finger protein 510 |
| chr11_-_125111708 | 0.67 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr3_-_132037800 | 0.65 |
ENST00000617767.4
|
CPNE4
|
copine 4 |
| chr17_-_41124178 | 0.65 |
ENST00000394014.2
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr18_-_63661884 | 0.64 |
ENST00000332821.8
ENST00000283752.10 |
SERPINB3
|
serpin family B member 3 |
| chr2_-_112433519 | 0.64 |
ENST00000496537.1
ENST00000330575.9 ENST00000302558.8 |
RGPD8
|
RANBP2 like and GRIP domain containing 8 |
| chr2_+_32277883 | 0.62 |
ENST00000238831.9
|
YIPF4
|
Yip1 domain family member 4 |
| chr3_+_141262614 | 0.62 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chrX_+_18425579 | 0.62 |
ENST00000379996.7
|
CDKL5
|
cyclin dependent kinase like 5 |
| chr15_-_44711306 | 0.62 |
ENST00000682850.1
|
PATL2
|
PAT1 homolog 2 |
| chr1_-_248277976 | 0.61 |
ENST00000641220.1
|
OR2T33
|
olfactory receptor family 2 subfamily T member 33 |
| chr3_+_148791058 | 0.61 |
ENST00000491148.5
|
CPB1
|
carboxypeptidase B1 |
| chr7_+_138460238 | 0.61 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr16_+_86566821 | 0.61 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr14_+_55027576 | 0.60 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr1_-_53940100 | 0.58 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr15_-_40874216 | 0.58 |
ENST00000220507.5
|
RHOV
|
ras homolog family member V |
| chr22_+_18150162 | 0.57 |
ENST00000215794.8
|
USP18
|
ubiquitin specific peptidase 18 |
| chr1_+_177170916 | 0.57 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chrX_-_6535118 | 0.57 |
ENST00000381089.7
ENST00000612369.4 ENST00000398729.1 |
VCX3A
|
variable charge X-linked 3A |
| chr14_-_105242605 | 0.56 |
ENST00000549655.5
|
BRF1
|
BRF1 RNA polymerase III transcription initiation factor subunit |
| chrX_-_53422170 | 0.56 |
ENST00000675504.1
|
SMC1A
|
structural maintenance of chromosomes 1A |
| chr6_+_27865308 | 0.56 |
ENST00000613174.2
|
H2AC16
|
H2A clustered histone 16 |
| chr19_-_14835252 | 0.55 |
ENST00000641666.1
ENST00000642030.1 ENST00000642000.1 |
OR7C1
|
olfactory receptor family 7 subfamily C member 1 |
| chr4_-_185956348 | 0.55 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_99219230 | 0.55 |
ENST00000394897.5
ENST00000508558.1 ENST00000394899.6 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr14_+_55027200 | 0.55 |
ENST00000395472.2
ENST00000555846.2 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr17_-_41184895 | 0.53 |
ENST00000620667.1
ENST00000398472.2 |
KRTAP4-1
|
keratin associated protein 4-1 |
| chr14_+_24120956 | 0.53 |
ENST00000558325.2
|
ENSG00000259371.2
|
novel protein |
| chr11_+_74592567 | 0.52 |
ENST00000263681.7
ENST00000527458.5 ENST00000532497.5 ENST00000530511.5 |
POLD3
|
DNA polymerase delta 3, accessory subunit |
| chr5_-_59276109 | 0.51 |
ENST00000503258.5
|
PDE4D
|
phosphodiesterase 4D |
| chr5_+_162067990 | 0.50 |
ENST00000641017.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr8_+_66775178 | 0.50 |
ENST00000396596.2
ENST00000521960.5 ENST00000522398.5 ENST00000522629.5 ENST00000520976.5 |
SGK3
|
serum/glucocorticoid regulated kinase family member 3 |
| chr5_+_162068031 | 0.49 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr11_+_112175526 | 0.49 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr16_+_31713231 | 0.47 |
ENST00000539915.5
ENST00000316491.13 ENST00000398696.3 ENST00000534369.1 ENST00000530881.5 ENST00000529515.1 |
ZNF720
|
zinc finger protein 720 |
| chr5_+_162067858 | 0.47 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr6_-_55875583 | 0.47 |
ENST00000370830.4
|
BMP5
|
bone morphogenetic protein 5 |
| chr9_-_5833014 | 0.46 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr8_+_103880412 | 0.46 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr13_+_48256214 | 0.45 |
ENST00000650237.1
|
ITM2B
|
integral membrane protein 2B |
| chr4_-_88697810 | 0.44 |
ENST00000323061.7
|
NAP1L5
|
nucleosome assembly protein 1 like 5 |
| chr11_+_72216774 | 0.43 |
ENST00000619261.4
ENST00000454954.6 ENST00000541003.5 ENST00000539412.5 ENST00000298223.11 ENST00000536778.5 ENST00000535625.5 ENST00000321324.11 |
FOLR2
|
folate receptor beta |
| chr10_-_27240505 | 0.43 |
ENST00000375888.5
ENST00000676732.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_+_112647059 | 0.42 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr12_+_57782742 | 0.42 |
ENST00000540550.6
ENST00000323833.12 ENST00000652027.2 ENST00000550559.5 ENST00000548851.5 ENST00000543727.5 ENST00000434359.5 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr1_-_170074568 | 0.42 |
ENST00000367767.5
ENST00000361580.7 ENST00000538366.5 |
KIFAP3
|
kinesin associated protein 3 |
| chr4_-_185956652 | 0.42 |
ENST00000355634.9
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_76072654 | 0.41 |
ENST00000369950.8
ENST00000611179.4 ENST00000369963.5 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr17_+_7407838 | 0.41 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr1_+_35268663 | 0.41 |
ENST00000314607.11
|
ZMYM4
|
zinc finger MYM-type containing 4 |
| chr10_+_89332484 | 0.40 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr3_+_38039199 | 0.40 |
ENST00000346219.7
ENST00000308059.11 |
DLEC1
|
DLEC1 cilia and flagella associated protein |
| chr12_+_9827517 | 0.40 |
ENST00000537723.5
|
KLRF1
|
killer cell lectin like receptor F1 |
| chr17_-_5035418 | 0.40 |
ENST00000254853.10
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 member 1 |
| chr11_+_2384603 | 0.39 |
ENST00000527343.5
ENST00000464784.6 |
CD81
|
CD81 molecule |
| chr6_+_110180418 | 0.39 |
ENST00000368930.5
ENST00000307731.2 |
CDC40
|
cell division cycle 40 |
| chr12_-_121039204 | 0.39 |
ENST00000620239.5
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr9_-_14314132 | 0.38 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B |
| chrY_-_6872608 | 0.37 |
ENST00000383036.1
|
AMELY
|
amelogenin Y-linked |
| chr1_-_219613069 | 0.37 |
ENST00000367211.6
ENST00000651890.1 |
ZC3H11B
|
zinc finger CCCH-type containing 11B |
| chr2_+_233917371 | 0.37 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr9_-_113303271 | 0.37 |
ENST00000297894.5
ENST00000489339.2 |
RNF183
|
ring finger protein 183 |
| chr4_+_26343156 | 0.36 |
ENST00000680928.1
ENST00000681260.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_+_93677556 | 0.36 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr1_-_114581589 | 0.35 |
ENST00000369541.4
|
BCAS2
|
BCAS2 pre-mRNA processing factor |
| chr11_+_59511368 | 0.35 |
ENST00000641278.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chr6_+_20534441 | 0.35 |
ENST00000274695.8
ENST00000613575.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1 like 1 |
| chr7_-_16465728 | 0.34 |
ENST00000307068.5
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr22_+_23856703 | 0.34 |
ENST00000345044.10
|
SLC2A11
|
solute carrier family 2 member 11 |
| chr12_-_121039156 | 0.32 |
ENST00000339275.10
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr10_-_122845850 | 0.32 |
ENST00000392790.6
|
CUZD1
|
CUB and zona pellucida like domains 1 |
| chr1_-_44986568 | 0.31 |
ENST00000372183.7
ENST00000360403.7 ENST00000620860.4 |
EIF2B3
|
eukaryotic translation initiation factor 2B subunit gamma |
| chr17_-_55511434 | 0.30 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36 |
| chr14_+_39175221 | 0.30 |
ENST00000556530.1
ENST00000216832.9 |
PNN
|
pinin, desmosome associated protein |
| chr5_-_56116946 | 0.28 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr6_+_132538290 | 0.28 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr4_-_186555567 | 0.28 |
ENST00000307161.5
|
MTNR1A
|
melatonin receptor 1A |
| chr1_+_152878312 | 0.28 |
ENST00000368765.4
|
SMCP
|
sperm mitochondria associated cysteine rich protein |
| chr5_+_141412979 | 0.27 |
ENST00000612503.1
ENST00000398610.3 |
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr4_-_88284616 | 0.27 |
ENST00000508256.5
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr4_-_88284590 | 0.26 |
ENST00000510548.6
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr8_+_97887903 | 0.26 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr12_-_121039236 | 0.26 |
ENST00000257570.9
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr15_-_78620964 | 0.26 |
ENST00000326828.6
|
CHRNA3
|
cholinergic receptor nicotinic alpha 3 subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 2.3 | 6.9 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 2.1 | 6.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.7 | 5.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) negative regulation of interleukin-6 biosynthetic process(GO:0045409) glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.6 | 1.8 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.5 | 1.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.5 | 1.8 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.4 | 8.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.4 | 4.1 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 4.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 3.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 0.9 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.3 | 2.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 2.0 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.3 | 0.8 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 6.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.3 | 0.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 1.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 4.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.2 | 0.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 1.9 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 0.6 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 2.8 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 4.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.2 | 1.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.2 | 0.9 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 2.4 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.2 | 0.5 | GO:0046247 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.2 | 12.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 0.5 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.2 | 1.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.4 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.7 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.6 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 0.4 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.1 | 2.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.7 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.3 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.4 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.4 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 1.9 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.6 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 1.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.4 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.1 | 0.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) tRNA transcription(GO:0009304) |
| 0.1 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 3.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.6 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 5.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.4 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 1.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 2.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.6 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 1.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.9 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.4 | GO:0043031 | T-helper 2 cell cytokine production(GO:0035745) negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.5 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 2.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.3 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 4.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 1.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 2.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.3 | 6.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 1.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.6 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 10.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 6.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 2.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 2.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 10.7 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 1.5 | 4.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.0 | 6.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.8 | 2.4 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.7 | 4.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.6 | 2.8 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.5 | 1.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 6.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 2.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.3 | 4.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.0 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 2.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 4.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.6 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.1 | 2.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 4.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 5.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.3 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 4.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.1 | 0.3 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.8 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.7 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 3.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 7.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 14.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 3.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 4.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 11.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 9.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 4.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 5.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 3.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |