Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for EN1_ESX1_GBX1
Z-value: 0.74
Transcription factors associated with EN1_ESX1_GBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
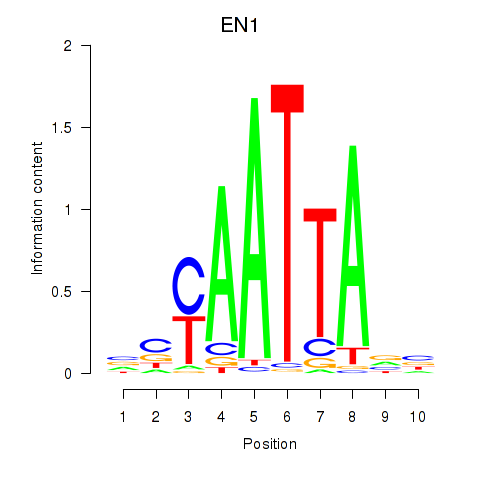
EN1
|
ENSG00000163064.7 | EN1 |
|
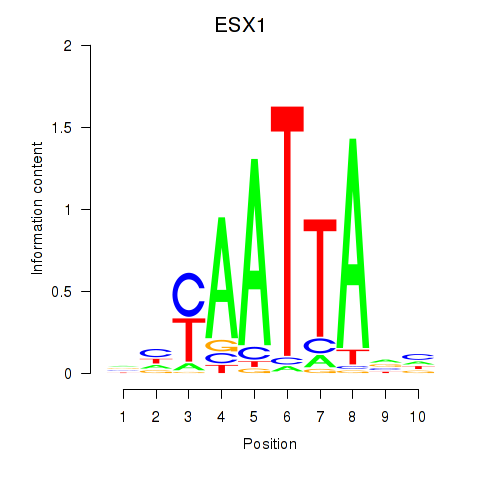
ESX1
|
ENSG00000123576.5 | ESX1 |
|
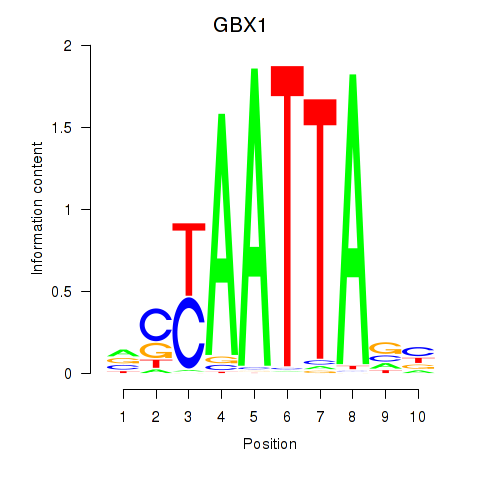
GBX1
|
ENSG00000164900.5 | GBX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESX1 | hg38_v1_chrX_-_104254921_104254936 | -0.14 | 4.7e-01 | Click! |
| EN1 | hg38_v1_chr2_-_118847638_118847654 | -0.13 | 4.9e-01 | Click! |
| GBX1 | hg38_v1_chr7_-_151167692_151167697 | 0.05 | 7.8e-01 | Click! |
Activity profile of EN1_ESX1_GBX1 motif
Sorted Z-values of EN1_ESX1_GBX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EN1_ESX1_GBX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_16978276 | 3.84 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr4_+_168092530 | 2.55 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr3_+_111999326 | 1.43 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr6_+_130018565 | 1.36 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr3_-_195811889 | 1.36 |
ENST00000475231.5
|
MUC4
|
mucin 4, cell surface associated |
| chr12_-_27970047 | 1.26 |
ENST00000395868.7
|
PTHLH
|
parathyroid hormone like hormone |
| chr3_-_74521140 | 1.18 |
ENST00000263665.6
|
CNTN3
|
contactin 3 |
| chr3_-_195811857 | 1.14 |
ENST00000349607.8
ENST00000346145.8 |
MUC4
|
mucin 4, cell surface associated |
| chr3_-_195811916 | 1.14 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated |
| chr12_-_27970273 | 1.00 |
ENST00000542963.1
ENST00000535992.5 |
PTHLH
|
parathyroid hormone like hormone |
| chr2_+_157257687 | 0.97 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr5_+_136059151 | 0.92 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr8_+_7881387 | 0.89 |
ENST00000314357.4
|
DEFB103A
|
defensin beta 103A |
| chr6_+_26199509 | 0.89 |
ENST00000356530.5
|
H2BC7
|
H2B clustered histone 7 |
| chr10_-_29736956 | 0.84 |
ENST00000674475.1
|
SVIL
|
supervillin |
| chr11_-_101129806 | 0.83 |
ENST00000325455.10
ENST00000617858.4 ENST00000619228.2 |
PGR
|
progesterone receptor |
| chr3_+_111998739 | 0.82 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr5_+_31193739 | 0.81 |
ENST00000514738.5
|
CDH6
|
cadherin 6 |
| chr3_+_111999189 | 0.78 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr19_+_44905785 | 0.77 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr5_+_151259793 | 0.76 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr3_-_142029108 | 0.76 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr16_-_55833085 | 0.74 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1 |
| chr1_-_74733253 | 0.72 |
ENST00000417775.5
|
CRYZ
|
crystallin zeta |
| chr3_+_111998915 | 0.70 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr12_+_26195313 | 0.70 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr3_+_122055355 | 0.68 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chrX_+_7219431 | 0.65 |
ENST00000674499.1
ENST00000217961.5 |
STS
|
steroid sulfatase |
| chr4_-_56681588 | 0.65 |
ENST00000554144.5
ENST00000381260.7 |
HOPX
|
HOP homeobox |
| chr3_-_138329839 | 0.63 |
ENST00000333911.9
ENST00000383180.6 |
NME9
|
NME/NM23 family member 9 |
| chr4_-_56681288 | 0.63 |
ENST00000556376.6
ENST00000420433.6 |
HOPX
|
HOP homeobox |
| chr1_+_209704836 | 0.63 |
ENST00000367027.5
|
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr17_-_40755328 | 0.62 |
ENST00000312150.5
|
KRT25
|
keratin 25 |
| chr5_+_31193678 | 0.61 |
ENST00000265071.3
|
CDH6
|
cadherin 6 |
| chr2_-_207167220 | 0.61 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr2_-_187554351 | 0.60 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr17_-_41055211 | 0.60 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr11_-_18236795 | 0.60 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr6_-_110179995 | 0.59 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr10_-_49762276 | 0.57 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr12_-_10802627 | 0.57 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7 |
| chr1_+_202348687 | 0.56 |
ENST00000608999.6
ENST00000391959.5 ENST00000480184.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr12_-_10826358 | 0.55 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr6_-_111759053 | 0.54 |
ENST00000462856.6
ENST00000229471.8 |
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr4_-_67883987 | 0.54 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr8_-_42377227 | 0.53 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr10_-_49762335 | 0.51 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr12_-_9869345 | 0.48 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr1_-_201171545 | 0.48 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr12_+_26195543 | 0.47 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr7_+_130344810 | 0.47 |
ENST00000497503.5
ENST00000463587.5 ENST00000461828.5 ENST00000474905.6 ENST00000494311.1 ENST00000466363.6 |
CPA5
|
carboxypeptidase A5 |
| chr14_+_21841182 | 0.46 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1 |
| chr6_-_39725387 | 0.45 |
ENST00000287152.12
|
KIF6
|
kinesin family member 6 |
| chr14_+_74348440 | 0.43 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr3_+_155080307 | 0.43 |
ENST00000360490.7
|
MME
|
membrane metalloendopeptidase |
| chr15_+_88621290 | 0.42 |
ENST00000332810.4
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr1_+_27934980 | 0.42 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr19_+_7030578 | 0.42 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr5_+_177384430 | 0.42 |
ENST00000512593.5
ENST00000324417.6 |
SLC34A1
|
solute carrier family 34 member 1 |
| chr20_-_37261808 | 0.42 |
ENST00000373614.7
|
GHRH
|
growth hormone releasing hormone |
| chr18_+_58221535 | 0.41 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr2_+_186694007 | 0.41 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr20_-_52105644 | 0.41 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr1_+_160190567 | 0.40 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr9_+_72577369 | 0.40 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr18_+_61333424 | 0.40 |
ENST00000262717.9
|
CDH20
|
cadherin 20 |
| chr1_+_27935110 | 0.40 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr11_-_101129706 | 0.40 |
ENST00000534013.5
|
PGR
|
progesterone receptor |
| chr15_-_41972504 | 0.39 |
ENST00000220325.9
|
EHD4
|
EH domain containing 4 |
| chr6_-_26199272 | 0.39 |
ENST00000650491.1
ENST00000635200.1 ENST00000341023.2 |
ENSG00000282988.2
H2AC7
|
novel protein H2A clustered histone 7 |
| chr14_+_22271921 | 0.38 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr7_-_101217569 | 0.38 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr12_-_21910853 | 0.37 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr12_+_107318395 | 0.37 |
ENST00000420571.6
ENST00000280758.10 |
BTBD11
|
BTB domain containing 11 |
| chr11_-_125111579 | 0.37 |
ENST00000532156.5
ENST00000532407.5 ENST00000279968.8 ENST00000527766.5 ENST00000529583.5 ENST00000524373.5 ENST00000527271.5 ENST00000526175.5 ENST00000529609.5 ENST00000682305.1 ENST00000533273.1 |
TMEM218
|
transmembrane protein 218 |
| chr6_-_127459364 | 0.37 |
ENST00000487331.2
ENST00000483725.8 |
KIAA0408
|
KIAA0408 |
| chr17_-_66229380 | 0.36 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr5_-_83673544 | 0.36 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr11_-_122116215 | 0.36 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr12_+_119668109 | 0.36 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr1_+_151060357 | 0.36 |
ENST00000368921.5
|
MLLT11
|
MLLT11 transcription factor 7 cofactor |
| chr20_-_57711536 | 0.36 |
ENST00000265626.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_+_131250375 | 0.36 |
ENST00000474850.2
|
AKAP7
|
A-kinase anchoring protein 7 |
| chrX_+_108045050 | 0.36 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_-_89352395 | 0.35 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr5_-_147906530 | 0.35 |
ENST00000318315.5
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr19_-_46634685 | 0.35 |
ENST00000300873.4
|
GNG8
|
G protein subunit gamma 8 |
| chr12_+_28257195 | 0.35 |
ENST00000381259.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr2_+_170178136 | 0.35 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chrX_-_18672101 | 0.35 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr15_-_55270280 | 0.35 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr14_-_106038355 | 0.34 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr12_-_47079926 | 0.34 |
ENST00000429635.1
ENST00000550413.2 |
AMIGO2
|
adhesion molecule with Ig like domain 2 |
| chr6_-_116545658 | 0.34 |
ENST00000368602.4
|
TRAPPC3L
|
trafficking protein particle complex 3 like |
| chrX_+_101550537 | 0.34 |
ENST00000372829.8
|
ARMCX1
|
armadillo repeat containing X-linked 1 |
| chr4_-_142305935 | 0.33 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_-_120054225 | 0.33 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr16_+_86566821 | 0.33 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr8_-_7385558 | 0.33 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr21_+_30396030 | 0.33 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chrX_+_108044967 | 0.33 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_-_10130241 | 0.32 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr17_-_59151794 | 0.32 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_+_33039996 | 0.32 |
ENST00000432887.5
ENST00000528898.1 ENST00000531632.6 |
TCP11L1
|
t-complex 11 like 1 |
| chr4_-_76007501 | 0.32 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr19_+_41363989 | 0.32 |
ENST00000413014.6
|
TMEM91
|
transmembrane protein 91 |
| chr16_-_29899245 | 0.32 |
ENST00000537485.5
|
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr11_+_59436469 | 0.32 |
ENST00000641045.1
|
OR5A1
|
olfactory receptor family 5 subfamily A member 1 |
| chr11_-_117876892 | 0.32 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr7_-_41703062 | 0.31 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr17_-_81683659 | 0.31 |
ENST00000574938.5
ENST00000570561.5 ENST00000573392.5 ENST00000576135.5 ENST00000573715.2 |
ARL16
|
ADP ribosylation factor like GTPase 16 |
| chr15_-_55270874 | 0.31 |
ENST00000567380.5
ENST00000565972.5 ENST00000569493.5 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_-_23347704 | 0.31 |
ENST00000619562.4
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr17_-_9791586 | 0.31 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chr3_-_125055987 | 0.31 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr11_-_117877463 | 0.31 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_-_10130143 | 0.31 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr9_+_107306459 | 0.31 |
ENST00000457811.1
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr4_-_108762964 | 0.31 |
ENST00000512646.5
ENST00000411864.6 ENST00000296486.8 ENST00000510706.5 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr13_-_36214584 | 0.30 |
ENST00000317764.6
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr14_+_22281097 | 0.30 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr7_+_130344837 | 0.30 |
ENST00000485477.5
ENST00000431780.6 |
CPA5
|
carboxypeptidase A5 |
| chr6_+_29099490 | 0.30 |
ENST00000641659.2
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr3_-_179266971 | 0.30 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr7_-_24980148 | 0.30 |
ENST00000313367.7
|
OSBPL3
|
oxysterol binding protein like 3 |
| chr19_-_3557563 | 0.30 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr19_-_7021431 | 0.30 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr1_-_93681829 | 0.30 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr6_+_26158115 | 0.30 |
ENST00000377777.5
ENST00000289316.2 |
H2BC5
|
H2B clustered histone 5 |
| chr10_+_47322450 | 0.30 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr11_+_75815180 | 0.30 |
ENST00000356136.8
|
UVRAG
|
UV radiation resistance associated |
| chr2_+_209579598 | 0.29 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr4_-_8871817 | 0.29 |
ENST00000400677.5
|
HMX1
|
H6 family homeobox 1 |
| chr3_-_151316795 | 0.29 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr3_-_187291882 | 0.29 |
ENST00000392470.6
ENST00000169293.10 ENST00000439271.1 ENST00000392472.6 ENST00000392475.2 |
MASP1
|
mannan binding lectin serine peptidase 1 |
| chr11_-_117876719 | 0.29 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_71821548 | 0.29 |
ENST00000525199.1
|
ZNF705E
|
zinc finger protein 705E |
| chr10_-_54801179 | 0.29 |
ENST00000373955.5
|
PCDH15
|
protocadherin related 15 |
| chr6_-_27912396 | 0.28 |
ENST00000303324.4
|
OR2B2
|
olfactory receptor family 2 subfamily B member 2 |
| chr10_-_48605032 | 0.28 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr12_-_119803383 | 0.27 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr8_-_7430348 | 0.27 |
ENST00000318124.3
|
DEFB103B
|
defensin beta 103B |
| chr12_-_89352487 | 0.27 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr8_+_12108172 | 0.27 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D |
| chr19_-_51019699 | 0.26 |
ENST00000358789.8
|
KLK10
|
kallikrein related peptidase 10 |
| chr15_+_21579912 | 0.26 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr13_-_30306997 | 0.26 |
ENST00000380617.7
ENST00000441394.1 |
KATNAL1
|
katanin catalytic subunit A1 like 1 |
| chr18_+_36544544 | 0.26 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr17_-_41047267 | 0.26 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr19_-_14529193 | 0.26 |
ENST00000596853.6
ENST00000676515.1 ENST00000678338.1 ENST00000595992.6 ENST00000677848.1 ENST00000677762.1 ENST00000678009.1 ENST00000596075.2 ENST00000601533.6 ENST00000396969.8 ENST00000598692.2 ENST00000678098.1 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr6_+_29170907 | 0.25 |
ENST00000641417.1
|
OR2J2
|
olfactory receptor family 2 subfamily J member 2 |
| chr21_+_29130630 | 0.25 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr5_+_136058849 | 0.25 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr18_-_14132423 | 0.25 |
ENST00000589498.5
ENST00000590202.3 |
ZNF519
|
zinc finger protein 519 |
| chr6_+_63521738 | 0.24 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr5_-_9630351 | 0.24 |
ENST00000382492.4
|
TAS2R1
|
taste 2 receptor member 1 |
| chr4_-_115113614 | 0.24 |
ENST00000264363.7
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr1_-_117929557 | 0.24 |
ENST00000369442.3
ENST00000369443.10 |
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr8_-_18887018 | 0.24 |
ENST00000523619.5
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr14_-_22957061 | 0.24 |
ENST00000557591.5
ENST00000541587.6 ENST00000490506.5 ENST00000554406.1 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr5_-_126595237 | 0.24 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr19_+_3762705 | 0.24 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr7_+_154305256 | 0.24 |
ENST00000619756.4
|
DPP6
|
dipeptidyl peptidase like 6 |
| chr7_-_73624492 | 0.23 |
ENST00000414749.6
ENST00000429400.6 ENST00000434326.5 ENST00000313375.8 ENST00000354613.5 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein like |
| chr16_-_20691256 | 0.23 |
ENST00000307493.8
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr8_+_7926337 | 0.23 |
ENST00000400120.3
|
ZNF705B
|
zinc finger protein 705B |
| chr10_-_97687191 | 0.23 |
ENST00000370626.4
|
AVPI1
|
arginine vasopressin induced 1 |
| chr6_+_29550407 | 0.23 |
ENST00000641137.1
|
OR2I1P
|
olfactory receptor family 2 subfamily I member 1 pseudogene |
| chr3_+_130850585 | 0.23 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr17_-_40994159 | 0.23 |
ENST00000391586.3
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr14_+_32329341 | 0.23 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr22_-_18936142 | 0.22 |
ENST00000438924.5
ENST00000457083.1 ENST00000357068.11 ENST00000420436.5 ENST00000334029.6 ENST00000610940.4 |
PRODH
|
proline dehydrogenase 1 |
| chr12_+_80716906 | 0.22 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr1_+_151762899 | 0.22 |
ENST00000635322.1
ENST00000321531.10 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr4_-_88284590 | 0.22 |
ENST00000510548.6
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr4_-_88284616 | 0.22 |
ENST00000508256.5
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr14_-_22957100 | 0.22 |
ENST00000555367.5
|
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr8_+_22567038 | 0.22 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr16_-_29899532 | 0.22 |
ENST00000308713.9
ENST00000617533.5 |
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr12_-_6124662 | 0.22 |
ENST00000261405.10
|
VWF
|
von Willebrand factor |
| chr10_-_13707536 | 0.22 |
ENST00000632570.1
ENST00000477221.2 |
FRMD4A
|
FERM domain containing 4A |
| chr17_-_81683782 | 0.22 |
ENST00000622299.5
|
ARL16
|
ADP ribosylation factor like GTPase 16 |
| chr14_-_22957128 | 0.22 |
ENST00000342454.12
ENST00000555986.5 ENST00000554516.5 ENST00000347758.6 ENST00000206474.11 ENST00000555040.5 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr20_+_58907981 | 0.22 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr10_-_48652493 | 0.22 |
ENST00000435790.6
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr11_+_65181194 | 0.21 |
ENST00000533820.5
|
CAPN1
|
calpain 1 |
| chr17_+_8002610 | 0.21 |
ENST00000254854.5
|
GUCY2D
|
guanylate cyclase 2D, retinal |
| chr5_+_127649018 | 0.21 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr12_-_10453330 | 0.21 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr8_-_130386864 | 0.21 |
ENST00000521426.5
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr7_+_124476371 | 0.21 |
ENST00000473520.1
|
SSU72P8
|
SSU72 pseudogene 8 |
| chr1_-_27672178 | 0.21 |
ENST00000339145.8
ENST00000361157.11 ENST00000362020.4 ENST00000679644.1 |
IFI6
|
interferon alpha inducible protein 6 |
| chr16_+_11965234 | 0.21 |
ENST00000562385.1
|
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr9_-_37025733 | 0.21 |
ENST00000651550.1
|
PAX5
|
paired box 5 |
| chr8_-_134510182 | 0.21 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr9_+_122371036 | 0.21 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr4_+_69931066 | 0.20 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr4_-_142305826 | 0.20 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr15_+_62561361 | 0.20 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chr16_+_24729692 | 0.20 |
ENST00000315183.11
|
TNRC6A
|
trinucleotide repeat containing adaptor 6A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 0.8 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 1.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 0.7 | GO:0002668 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.2 | 4.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 0.7 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.1 | 0.4 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.4 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.6 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.5 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.1 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 0.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.1 | 0.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.2 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.1 | 2.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.2 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0015855 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.7 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) mast cell proliferation(GO:0070662) |
| 0.0 | 0.5 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.7 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.3 | GO:0097680 | maintenance of Golgi location(GO:0051684) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.9 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0071626 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.0 | 1.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.3 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.9 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0060770 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0018026 | histone H3-K36 methylation(GO:0010452) peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.0 | 0.5 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 0.8 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 3.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 3.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.0 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.3 | 0.8 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.2 | 0.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 0.7 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 1.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 0.7 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.2 | 0.8 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.7 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.4 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.3 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.1 | 0.4 | GO:0050211 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.5 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.2 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 2.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.3 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 4.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 4.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 5.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 3.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.0 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |