Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
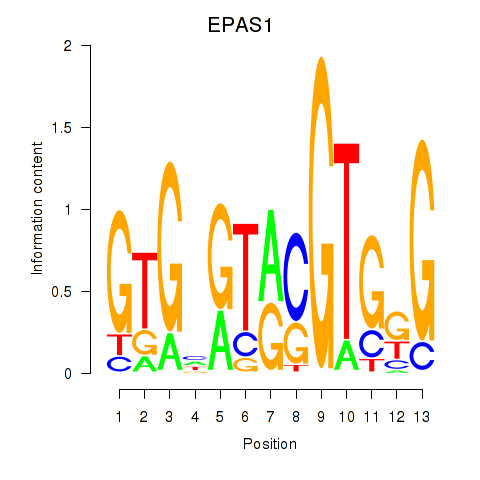
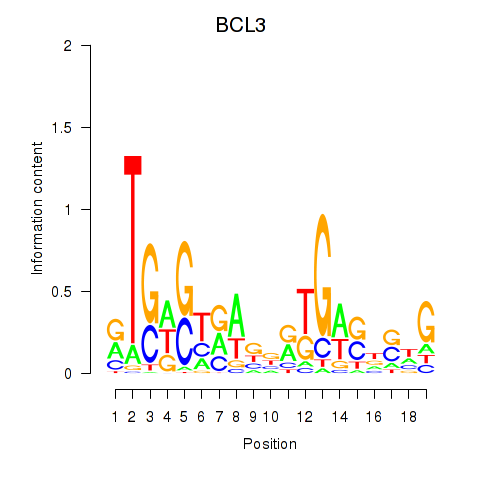
Results for EPAS1_BCL3
Z-value: 1.27
Transcription factors associated with EPAS1_BCL3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EPAS1
|
ENSG00000116016.14 | EPAS1 |
|
BCL3
|
ENSG00000069399.15 | BCL3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EPAS1 | hg38_v1_chr2_+_46297397_46297414 | -0.23 | 2.2e-01 | Click! |
| BCL3 | hg38_v1_chr19_+_44751251_44751321 | -0.18 | 3.4e-01 | Click! |
Activity profile of EPAS1_BCL3 motif
Sorted Z-values of EPAS1_BCL3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EPAS1_BCL3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_40351026 | 1.92 |
ENST00000448599.2
|
PHGR1
|
proline, histidine and glycine rich 1 |
| chr7_+_154305256 | 1.89 |
ENST00000619756.4
|
DPP6
|
dipeptidyl peptidase like 6 |
| chr12_+_56021316 | 1.67 |
ENST00000547791.2
|
IKZF4
|
IKAROS family zinc finger 4 |
| chr3_-_116444983 | 1.43 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr20_-_2800576 | 1.39 |
ENST00000380605.3
|
CPXM1
|
carboxypeptidase X, M14 family member 1 |
| chr20_-_45115149 | 1.38 |
ENST00000307971.7
ENST00000372789.5 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr16_+_82626955 | 1.34 |
ENST00000268613.14
ENST00000567109.6 ENST00000565636.5 ENST00000431540.7 ENST00000428848.7 |
CDH13
|
cadherin 13 |
| chr1_+_158254414 | 1.28 |
ENST00000289429.6
|
CD1A
|
CD1a molecule |
| chr19_+_35138778 | 1.22 |
ENST00000351325.9
ENST00000586871.5 ENST00000592174.1 |
FXYD1
ENSG00000221857.7
|
FXYD domain containing ion transport regulator 1 novel transcript |
| chr1_+_152514474 | 1.19 |
ENST00000368790.4
|
CRCT1
|
cysteine rich C-terminal 1 |
| chr2_+_237566574 | 1.18 |
ENST00000165524.1
|
PRLH
|
prolactin releasing hormone |
| chr17_+_21376321 | 1.16 |
ENST00000583088.6
|
KCNJ12
|
potassium inwardly rectifying channel subfamily J member 12 |
| chr6_-_6007511 | 1.05 |
ENST00000616243.1
|
NRN1
|
neuritin 1 |
| chr20_+_44715360 | 1.03 |
ENST00000190983.5
|
CCN5
|
cellular communication network factor 5 |
| chr10_-_97185758 | 1.00 |
ENST00000371070.8
|
SLIT1
|
slit guidance ligand 1 |
| chr3_-_187291882 | 0.98 |
ENST00000392470.6
ENST00000169293.10 ENST00000439271.1 ENST00000392472.6 ENST00000392475.2 |
MASP1
|
mannan binding lectin serine peptidase 1 |
| chr16_-_11273610 | 0.97 |
ENST00000327157.4
|
PRM3
|
protamine 3 |
| chr16_+_1528674 | 0.96 |
ENST00000253934.9
|
TMEM204
|
transmembrane protein 204 |
| chr9_+_34652167 | 0.95 |
ENST00000441545.7
ENST00000553620.5 |
IL11RA
|
interleukin 11 receptor subunit alpha |
| chr12_-_2835018 | 0.94 |
ENST00000337508.9
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chrX_+_1268807 | 0.94 |
ENST00000381524.8
ENST00000381529.9 ENST00000412290.6 |
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha |
| chr7_+_148339452 | 0.90 |
ENST00000463592.3
|
CNTNAP2
|
contactin associated protein 2 |
| chr16_+_4958289 | 0.89 |
ENST00000251170.12
|
SEC14L5
|
SEC14 like lipid binding 5 |
| chr16_-_20669855 | 0.88 |
ENST00000524149.5
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr1_-_39691450 | 0.87 |
ENST00000612703.3
ENST00000617690.2 |
HPCAL4
|
hippocalcin like 4 |
| chr20_+_44714835 | 0.86 |
ENST00000372868.6
|
CCN5
|
cellular communication network factor 5 |
| chr14_+_32934383 | 0.86 |
ENST00000551634.6
|
NPAS3
|
neuronal PAS domain protein 3 |
| chr8_-_69834970 | 0.85 |
ENST00000260126.9
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr14_-_94509469 | 0.84 |
ENST00000677451.1
|
SERPINA12
|
serpin family A member 12 |
| chr19_-_8981342 | 0.83 |
ENST00000397910.8
|
MUC16
|
mucin 16, cell surface associated |
| chr12_+_26195647 | 0.81 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr8_-_139704091 | 0.80 |
ENST00000648164.1
|
KCNK9
|
potassium two pore domain channel subfamily K member 9 |
| chr11_+_65833944 | 0.77 |
ENST00000308342.7
|
SNX32
|
sorting nexin 32 |
| chr19_+_35138993 | 0.77 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr7_-_36724380 | 0.75 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr8_+_27491125 | 0.75 |
ENST00000517536.5
ENST00000521400.6 ENST00000521780.5 ENST00000380476.7 ENST00000518379.5 ENST00000521684.1 |
EPHX2
|
epoxide hydrolase 2 |
| chr20_+_44714853 | 0.75 |
ENST00000372865.4
|
CCN5
|
cellular communication network factor 5 |
| chrX_+_1268786 | 0.74 |
ENST00000501036.7
ENST00000417535.7 |
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha |
| chr16_+_72056153 | 0.73 |
ENST00000576168.6
ENST00000567185.7 ENST00000567612.2 |
HP
|
haptoglobin |
| chr3_-_128493173 | 0.72 |
ENST00000498200.1
ENST00000341105.7 |
GATA2
|
GATA binding protein 2 |
| chr11_+_10450627 | 0.70 |
ENST00000396554.7
ENST00000524866.5 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr7_-_38368169 | 0.69 |
ENST00000390348.2
|
TRGV1
|
T cell receptor gamma variable 1 (non-functional) |
| chr7_-_76409682 | 0.69 |
ENST00000275560.4
|
SSC4D
|
scavenger receptor cysteine rich family member with 4 domains |
| chr19_-_48513161 | 0.68 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr10_-_133336862 | 0.66 |
ENST00000368555.3
ENST00000252939.9 ENST00000368558.1 |
CALY
|
calcyon neuron specific vesicular protein |
| chr8_+_76683779 | 0.64 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr3_-_127823177 | 0.64 |
ENST00000434178.6
|
MGLL
|
monoglyceride lipase |
| chr11_-_75351609 | 0.63 |
ENST00000420843.7
|
ARRB1
|
arrestin beta 1 |
| chr7_-_36724457 | 0.63 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr19_+_35139724 | 0.62 |
ENST00000588715.5
ENST00000588607.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr3_-_127823235 | 0.62 |
ENST00000398104.5
|
MGLL
|
monoglyceride lipase |
| chr9_+_87498491 | 0.62 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1 |
| chr6_+_146027662 | 0.62 |
ENST00000492807.6
ENST00000361719.6 |
GRM1
|
glutamate metabotropic receptor 1 |
| chr6_-_152563271 | 0.61 |
ENST00000535896.7
ENST00000672122.1 |
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr3_+_127629161 | 0.61 |
ENST00000342480.7
|
PODXL2
|
podocalyxin like 2 |
| chr9_+_87497852 | 0.60 |
ENST00000408954.8
|
DAPK1
|
death associated protein kinase 1 |
| chr2_-_27119099 | 0.60 |
ENST00000402550.5
ENST00000402394.6 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr2_+_219572304 | 0.60 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha |
| chr3_-_127822835 | 0.60 |
ENST00000453507.6
|
MGLL
|
monoglyceride lipase |
| chr19_+_35139440 | 0.59 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr11_+_10450289 | 0.59 |
ENST00000444303.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr14_+_41608344 | 0.58 |
ENST00000554120.5
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr2_+_209579429 | 0.58 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2 |
| chr7_-_38330935 | 0.58 |
ENST00000390343.2
|
TRGV8
|
T cell receptor gamma variable 8 |
| chr10_-_5499544 | 0.58 |
ENST00000380332.5
|
CALML5
|
calmodulin like 5 |
| chr19_+_30372364 | 0.58 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536 |
| chr15_-_65422894 | 0.56 |
ENST00000352385.3
|
IGDCC4
|
immunoglobulin superfamily DCC subclass member 4 |
| chr4_-_16084002 | 0.55 |
ENST00000447510.7
|
PROM1
|
prominin 1 |
| chr9_+_87497222 | 0.54 |
ENST00000358077.9
|
DAPK1
|
death associated protein kinase 1 |
| chr1_+_109656092 | 0.54 |
ENST00000369836.9
ENST00000326729.9 |
GSTM4
|
glutathione S-transferase mu 4 |
| chr3_-_193554952 | 0.54 |
ENST00000392443.7
|
ATP13A4
|
ATPase 13A4 |
| chr18_-_23662810 | 0.54 |
ENST00000322980.13
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr19_+_37371092 | 0.53 |
ENST00000436120.7
|
ZNF527
|
zinc finger protein 527 |
| chr1_-_39691393 | 0.53 |
ENST00000372844.8
|
HPCAL4
|
hippocalcin like 4 |
| chr15_+_47717344 | 0.53 |
ENST00000558816.5
ENST00000536845.7 |
SEMA6D
|
semaphorin 6D |
| chr1_-_26360050 | 0.52 |
ENST00000475866.3
|
CRYBG2
|
crystallin beta-gamma domain containing 2 |
| chr2_-_85418421 | 0.52 |
ENST00000409275.1
|
CAPG
|
capping actin protein, gelsolin like |
| chr2_+_30231524 | 0.51 |
ENST00000395323.9
ENST00000406087.5 ENST00000404397.5 |
LBH
|
LBH regulator of WNT signaling pathway |
| chr9_+_18474100 | 0.50 |
ENST00000327883.11
ENST00000431052.6 ENST00000380570.8 ENST00000380548.9 |
ADAMTSL1
|
ADAMTS like 1 |
| chr1_+_153678680 | 0.50 |
ENST00000368680.4
|
NPR1
|
natriuretic peptide receptor 1 |
| chr15_+_74173693 | 0.50 |
ENST00000249842.8
|
ISLR
|
immunoglobulin superfamily containing leucine rich repeat |
| chr22_-_37484505 | 0.50 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_-_185152974 | 0.50 |
ENST00000335012.3
|
C3orf70
|
chromosome 3 open reading frame 70 |
| chr1_+_109656322 | 0.49 |
ENST00000369833.5
ENST00000336075.6 ENST00000638994.1 |
GSTM4
|
glutathione S-transferase mu 4 |
| chr22_+_39456996 | 0.49 |
ENST00000341184.7
|
MGAT3
|
beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase |
| chr3_+_134795248 | 0.49 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1 |
| chr10_+_100462969 | 0.48 |
ENST00000343737.6
|
WNT8B
|
Wnt family member 8B |
| chr3_+_113211539 | 0.48 |
ENST00000682979.1
ENST00000485230.5 |
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr7_-_132576463 | 0.48 |
ENST00000423507.6
|
PLXNA4
|
plexin A4 |
| chr13_-_98752619 | 0.48 |
ENST00000376503.10
|
SLC15A1
|
solute carrier family 15 member 1 |
| chr17_-_43545707 | 0.48 |
ENST00000545089.5
|
ETV4
|
ETS variant transcription factor 4 |
| chr8_+_109086585 | 0.48 |
ENST00000518632.2
|
TRHR
|
thyrotropin releasing hormone receptor |
| chr7_-_38349972 | 0.47 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr22_-_30574572 | 0.47 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_-_21345531 | 0.47 |
ENST00000649812.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr19_-_35134861 | 0.47 |
ENST00000591633.2
|
LGI4
|
leucine rich repeat LGI family member 4 |
| chr21_-_29939912 | 0.47 |
ENST00000399907.5
ENST00000399909.5 |
GRIK1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr19_+_56404314 | 0.46 |
ENST00000333201.13
ENST00000391778.3 |
ZNF583
|
zinc finger protein 583 |
| chr19_+_53521245 | 0.46 |
ENST00000649326.1
|
ZNF331
|
zinc finger protein 331 |
| chr1_+_203626775 | 0.46 |
ENST00000367218.7
|
ATP2B4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chrX_-_20116871 | 0.46 |
ENST00000379651.7
ENST00000443379.7 ENST00000379643.10 |
MAP7D2
|
MAP7 domain containing 2 |
| chr17_-_43546323 | 0.46 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr17_-_7207245 | 0.45 |
ENST00000649971.1
|
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr1_+_218346235 | 0.45 |
ENST00000366929.4
|
TGFB2
|
transforming growth factor beta 2 |
| chr8_+_10672623 | 0.45 |
ENST00000304519.10
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr3_-_193554885 | 0.45 |
ENST00000342695.9
|
ATP13A4
|
ATPase 13A4 |
| chr14_+_104724221 | 0.45 |
ENST00000330877.7
|
ADSS1
|
adenylosuccinate synthase 1 |
| chr8_+_22057857 | 0.44 |
ENST00000517305.4
ENST00000265800.9 ENST00000517418.5 |
DMTN
|
dematin actin binding protein |
| chr17_-_7114240 | 0.43 |
ENST00000446679.6
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr17_+_8039106 | 0.43 |
ENST00000573359.1
|
ALOX15B
|
arachidonate 15-lipoxygenase type B |
| chr11_+_117986386 | 0.43 |
ENST00000227752.8
|
IL10RA
|
interleukin 10 receptor subunit alpha |
| chr9_+_72577939 | 0.43 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr18_+_63777773 | 0.43 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr1_-_44031352 | 0.43 |
ENST00000372306.7
ENST00000475075.6 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr7_+_74773962 | 0.43 |
ENST00000289473.10
|
NCF1
|
neutrophil cytosolic factor 1 |
| chr5_-_11588842 | 0.43 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chr9_-_14910421 | 0.43 |
ENST00000380880.4
|
FREM1
|
FRAS1 related extracellular matrix 1 |
| chr19_+_37594830 | 0.43 |
ENST00000589117.5
|
ZNF540
|
zinc finger protein 540 |
| chr14_+_70641896 | 0.43 |
ENST00000256367.3
|
TTC9
|
tetratricopeptide repeat domain 9 |
| chr12_+_120650492 | 0.43 |
ENST00000351200.6
|
CABP1
|
calcium binding protein 1 |
| chr1_-_23424692 | 0.42 |
ENST00000374601.7
ENST00000450454.7 |
TCEA3
|
transcription elongation factor A3 |
| chr18_-_23662868 | 0.42 |
ENST00000586087.1
ENST00000592179.6 |
ANKRD29
|
ankyrin repeat domain 29 |
| chr14_+_96039328 | 0.42 |
ENST00000553764.1
ENST00000555004.3 ENST00000556728.1 ENST00000553782.1 |
C14orf132
|
chromosome 14 open reading frame 132 |
| chrX_+_130171903 | 0.42 |
ENST00000257017.5
|
RAB33A
|
RAB33A, member RAS oncogene family |
| chr12_-_7665897 | 0.42 |
ENST00000229304.5
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr2_+_209579399 | 0.42 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr17_-_76027296 | 0.42 |
ENST00000301607.8
|
EVPL
|
envoplakin |
| chr9_-_114930508 | 0.41 |
ENST00000223795.3
ENST00000618336.4 |
TNFSF8
|
TNF superfamily member 8 |
| chr8_+_144107733 | 0.41 |
ENST00000323662.9
|
WDR97
|
WD repeat domain 97 |
| chr19_-_42937201 | 0.41 |
ENST00000406070.7
|
PSG7
|
pregnancy specific beta-1-glycoprotein 7 |
| chr19_+_55857437 | 0.41 |
ENST00000587891.5
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chrX_-_75523011 | 0.40 |
ENST00000373367.8
|
ZDHHC15
|
zinc finger DHHC-type palmitoyltransferase 15 |
| chr17_-_74859863 | 0.40 |
ENST00000293190.10
|
GRIN2C
|
glutamate ionotropic receptor NMDA type subunit 2C |
| chr8_+_62248591 | 0.40 |
ENST00000519049.6
|
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr2_+_209579598 | 0.40 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr2_-_27119012 | 0.39 |
ENST00000312734.8
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr9_+_72628020 | 0.39 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr9_+_87497675 | 0.39 |
ENST00000472284.5
ENST00000469640.6 |
DAPK1
|
death associated protein kinase 1 |
| chr14_+_79279403 | 0.39 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr5_-_148654130 | 0.39 |
ENST00000360693.7
|
HTR4
|
5-hydroxytryptamine receptor 4 |
| chr5_+_167754918 | 0.38 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chr16_+_66603874 | 0.38 |
ENST00000563672.5
ENST00000424011.6 |
CMTM3
|
CKLF like MARVEL transmembrane domain containing 3 |
| chr11_-_111912871 | 0.38 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr7_+_154305105 | 0.37 |
ENST00000332007.7
|
DPP6
|
dipeptidyl peptidase like 6 |
| chr8_+_68330923 | 0.37 |
ENST00000518698.6
|
C8orf34
|
chromosome 8 open reading frame 34 |
| chr1_-_44031446 | 0.37 |
ENST00000372310.8
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr16_+_50693568 | 0.37 |
ENST00000647318.2
ENST00000531674.1 |
NOD2
|
nucleotide binding oligomerization domain containing 2 |
| chr9_+_92974476 | 0.37 |
ENST00000337352.10
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr1_+_13584262 | 0.37 |
ENST00000376061.8
ENST00000513143.5 |
PDPN
|
podoplanin |
| chr2_+_74002685 | 0.37 |
ENST00000305799.8
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr1_+_13583762 | 0.37 |
ENST00000376057.8
ENST00000621990.5 ENST00000510906.5 |
PDPN
|
podoplanin |
| chr4_+_157220691 | 0.37 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr9_+_101028721 | 0.36 |
ENST00000374874.8
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr19_-_50511146 | 0.36 |
ENST00000594350.1
ENST00000601423.5 |
JOSD2
|
Josephin domain containing 2 |
| chr16_+_71626149 | 0.36 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr4_-_108167784 | 0.36 |
ENST00000438313.6
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chr19_-_9792991 | 0.36 |
ENST00000592587.1
|
ZNF846
|
zinc finger protein 846 |
| chr20_+_58689124 | 0.36 |
ENST00000525967.5
ENST00000525817.5 |
NPEPL1
|
aminopeptidase like 1 |
| chr19_+_35292145 | 0.36 |
ENST00000595791.5
ENST00000597035.5 ENST00000537831.2 ENST00000392213.8 |
MAG
|
myelin associated glycoprotein |
| chr2_+_33476640 | 0.36 |
ENST00000425210.5
ENST00000444784.5 ENST00000423159.5 ENST00000403687.8 |
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr3_-_127822455 | 0.36 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chrX_+_102651366 | 0.36 |
ENST00000415986.5
ENST00000444152.5 ENST00000361600.9 |
GPRASP1
|
G protein-coupled receptor associated sorting protein 1 |
| chr7_-_132576493 | 0.35 |
ENST00000321063.8
|
PLXNA4
|
plexin A4 |
| chr21_-_29939532 | 0.35 |
ENST00000327783.8
ENST00000389124.6 ENST00000389125.7 ENST00000399913.5 |
GRIK1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr17_-_7204502 | 0.35 |
ENST00000486626.8
ENST00000648263.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr19_-_11577632 | 0.35 |
ENST00000590420.1
ENST00000648477.1 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr2_-_219571529 | 0.35 |
ENST00000404537.6
|
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr12_-_23949642 | 0.35 |
ENST00000537393.5
ENST00000451604.7 ENST00000381381.6 |
SOX5
|
SRY-box transcription factor 5 |
| chr19_-_51723968 | 0.35 |
ENST00000222115.5
ENST00000540069.7 |
HAS1
|
hyaluronan synthase 1 |
| chr8_-_133297092 | 0.35 |
ENST00000522890.5
ENST00000675983.1 ENST00000518176.5 ENST00000323851.13 ENST00000522476.5 ENST00000518066.5 ENST00000521544.5 ENST00000674605.1 ENST00000518480.5 ENST00000523892.5 |
NDRG1
|
N-myc downstream regulated 1 |
| chr4_+_40196907 | 0.35 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr12_+_50057548 | 0.35 |
ENST00000228468.8
ENST00000447966.7 |
ASIC1
|
acid sensing ion channel subunit 1 |
| chrX_+_102651476 | 0.34 |
ENST00000537097.2
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1 |
| chr18_+_65751000 | 0.34 |
ENST00000397968.4
|
CDH7
|
cadherin 7 |
| chr1_+_20290869 | 0.34 |
ENST00000289815.13
ENST00000375079.6 |
VWA5B1
|
von Willebrand factor A domain containing 5B1 |
| chr4_-_56681588 | 0.34 |
ENST00000554144.5
ENST00000381260.7 |
HOPX
|
HOP homeobox |
| chr2_-_2331336 | 0.34 |
ENST00000648933.1
ENST00000644820.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr16_+_66604100 | 0.33 |
ENST00000562357.5
ENST00000562707.5 ENST00000361909.8 ENST00000460097.5 ENST00000565666.5 |
CMTM3
|
CKLF like MARVEL transmembrane domain containing 3 |
| chr17_+_1279655 | 0.33 |
ENST00000333813.4
|
TRARG1
|
trafficking regulator of GLUT4 (SLC2A4) 1 |
| chr16_+_981762 | 0.33 |
ENST00000293894.4
|
SOX8
|
SRY-box transcription factor 8 |
| chr15_-_64046322 | 0.33 |
ENST00000457488.5
ENST00000612884.4 |
DAPK2
|
death associated protein kinase 2 |
| chr2_-_27118900 | 0.33 |
ENST00000405600.5
ENST00000260595.9 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr7_+_1709162 | 0.33 |
ENST00000561626.3
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr9_-_14722725 | 0.33 |
ENST00000380911.4
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr1_+_228165794 | 0.32 |
ENST00000366711.4
|
IBA57
|
iron-sulfur cluster assembly factor IBA57 |
| chr4_+_157220654 | 0.32 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr3_-_184017863 | 0.32 |
ENST00000427120.6
ENST00000334444.11 ENST00000392579.6 ENST00000382494.6 ENST00000265586.10 ENST00000446941.2 |
ABCC5
|
ATP binding cassette subfamily C member 5 |
| chr14_-_24429665 | 0.32 |
ENST00000267406.11
|
CBLN3
|
cerebellin 3 precursor |
| chr4_+_40197023 | 0.32 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr16_+_4371840 | 0.32 |
ENST00000304735.4
|
VASN
|
vasorin |
| chr9_-_127877665 | 0.32 |
ENST00000644144.2
|
AK1
|
adenylate kinase 1 |
| chr2_-_218831791 | 0.32 |
ENST00000439262.6
ENST00000430489.1 |
PRKAG3
|
protein kinase AMP-activated non-catalytic subunit gamma 3 |
| chr7_+_142111739 | 0.32 |
ENST00000550469.6
ENST00000477922.3 |
MGAM2
|
maltase-glucoamylase 2 (putative) |
| chr7_+_101127095 | 0.32 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1 |
| chr10_+_110207587 | 0.32 |
ENST00000332674.9
ENST00000453116.5 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr19_-_50511203 | 0.31 |
ENST00000595669.5
|
JOSD2
|
Josephin domain containing 2 |
| chr5_+_146338835 | 0.31 |
ENST00000646991.2
|
POU4F3
|
POU class 4 homeobox 3 |
| chr8_-_88327475 | 0.31 |
ENST00000286614.11
|
MMP16
|
matrix metallopeptidase 16 |
| chr22_+_22818994 | 0.31 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 |
| chr8_-_72075575 | 0.31 |
ENST00000262209.5
|
TRPA1
|
transient receptor potential cation channel subfamily A member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.4 | 1.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.3 | 1.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.3 | 0.9 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 0.8 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.3 | 0.8 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.2 | 2.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.7 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 | 0.7 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.2 | 1.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.3 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.2 | 0.6 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.2 | 0.6 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.2 | 0.9 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 0.7 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.2 | 0.8 | GO:1901205 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) |
| 0.2 | 0.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.9 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.2 | 0.5 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.1 | 0.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.7 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 1.3 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 1.5 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.4 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.4 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.4 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.1 | 0.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.5 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 | 0.8 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.7 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.1 | 1.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.4 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 2.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.1 | 0.4 | GO:0060584 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.5 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 2.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.4 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.1 | 0.5 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.4 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.1 | 0.3 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.8 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.4 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.6 | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 0.2 | GO:1905237 | response to cyclosporin A(GO:1905237) positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.3 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.2 | GO:1903004 | regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.1 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.1 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.2 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.1 | 0.2 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.6 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.9 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.2 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.8 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.3 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.2 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.2 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.5 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.2 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.2 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.2 | GO:1904327 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.4 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.6 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.2 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.0 | 0.2 | GO:0031022 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.2 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 2.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 1.0 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.3 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.3 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.4 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.1 | GO:0061047 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.4 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.2 | GO:0003183 | mitral valve morphogenesis(GO:0003183) |
| 0.0 | 0.4 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.0 | 0.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.3 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.2 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.0 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.0 | 0.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0051792 | short-chain fatty acid biosynthetic process(GO:0051790) medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.4 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.4 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.7 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 2.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0060763 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 1.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.8 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 1.0 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.5 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.4 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 1.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 1.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0021930 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 1.2 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.3 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.6 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.7 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 0.6 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 3.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.7 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.4 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.5 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 3.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 3.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.4 | 1.3 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 0.7 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 0.9 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 0.5 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 1.3 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.4 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.3 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 0.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 2.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.4 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.1 | 0.5 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 1.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.3 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 2.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 2.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 1.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 1.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.6 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0035925 | C-C chemokine binding(GO:0019957) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 2.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 3.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |