Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for EVX2
Z-value: 0.26
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | EVX2 |
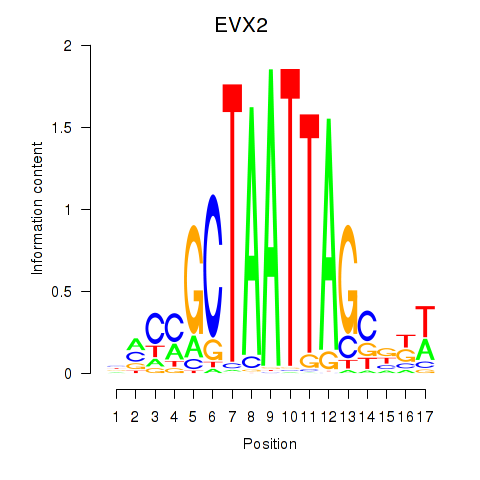
Activity profile of EVX2 motif
Sorted Z-values of EVX2 motif
Network of associatons between targets according to the STRING database.
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_111999326 | 0.66 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr11_-_115504389 | 0.66 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr3_+_111998915 | 0.54 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr3_+_111998739 | 0.54 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr3_+_111999189 | 0.53 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr1_+_27934980 | 0.25 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr1_+_27935110 | 0.23 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr20_-_51802433 | 0.22 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr5_-_97142579 | 0.21 |
ENST00000274382.9
|
LIX1
|
limb and CNS expressed 1 |
| chr20_-_51802509 | 0.18 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr4_+_143433491 | 0.14 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr4_+_76435216 | 0.10 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr4_-_73620629 | 0.09 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr4_+_112860912 | 0.09 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112861053 | 0.09 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112860981 | 0.08 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr2_+_71276553 | 0.08 |
ENST00000494621.5
ENST00000466975.5 ENST00000410075.5 ENST00000466330.5 |
ZNF638
|
zinc finger protein 638 |
| chr4_-_145180496 | 0.05 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4 |
| chr1_-_99766620 | 0.05 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr1_+_27935022 | 0.05 |
ENST00000411604.5
ENST00000373888.8 |
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr6_+_39792298 | 0.04 |
ENST00000633794.1
ENST00000274867.9 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr21_-_31160904 | 0.04 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr12_+_119668109 | 0.04 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr20_+_31879798 | 0.03 |
ENST00000620043.4
|
TTLL9
|
tubulin tyrosine ligase like 9 |
| chr6_-_62286161 | 0.03 |
ENST00000281156.5
|
KHDRBS2
|
KH RNA binding domain containing, signal transduction associated 2 |
| chr12_-_53336342 | 0.02 |
ENST00000537210.2
ENST00000536324.4 |
SP7
|
Sp7 transcription factor |
| chr17_+_18183803 | 0.01 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr1_-_151459169 | 0.01 |
ENST00000368863.6
ENST00000409503.5 ENST00000491586.5 ENST00000533351.5 |
POGZ
|
pogo transposable element derived with ZNF domain |
| chr12_+_28257195 | 0.01 |
ENST00000381259.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chrX_-_63755187 | 0.00 |
ENST00000635729.1
ENST00000623566.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr19_-_46634685 | 0.00 |
ENST00000300873.4
|
GNG8
|
G protein subunit gamma 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |