Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for FOSB
Z-value: 0.55
Transcription factors associated with FOSB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSB
|
ENSG00000125740.14 | FOSB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSB | hg38_v1_chr19_+_45469841_45469913 | 0.39 | 3.5e-02 | Click! |
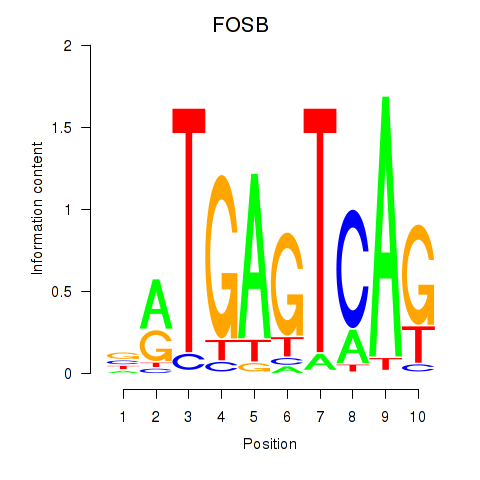
Activity profile of FOSB motif
Sorted Z-values of FOSB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_152908538 | 2.00 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr18_+_23873000 | 1.87 |
ENST00000269217.11
ENST00000587184.5 |
LAMA3
|
laminin subunit alpha 3 |
| chr1_-_153057504 | 1.78 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A |
| chr19_-_35528221 | 1.43 |
ENST00000588674.5
ENST00000452271.7 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_-_153094521 | 1.26 |
ENST00000368750.8
|
SPRR2E
|
small proline rich protein 2E |
| chr17_-_41612757 | 1.21 |
ENST00000301653.9
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr11_-_65900375 | 1.18 |
ENST00000312562.7
|
FOSL1
|
FOS like 1, AP-1 transcription factor subunit |
| chr6_+_73695779 | 1.18 |
ENST00000422508.6
ENST00000437994.6 |
CD109
|
CD109 molecule |
| chr6_+_73696145 | 1.14 |
ENST00000287097.6
|
CD109
|
CD109 molecule |
| chr1_-_153113507 | 1.13 |
ENST00000468739.2
|
SPRR2F
|
small proline rich protein 2F |
| chr1_-_153549238 | 1.08 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chr12_+_53098846 | 1.04 |
ENST00000650247.1
ENST00000549628.1 |
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr11_-_65900413 | 0.99 |
ENST00000448083.6
ENST00000531493.5 ENST00000532401.1 |
FOSL1
|
FOS like 1, AP-1 transcription factor subunit |
| chr1_-_153041111 | 0.97 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr3_-_48595267 | 0.97 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr1_-_153549120 | 0.97 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr22_+_31082860 | 0.90 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chr12_-_95116967 | 0.90 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr19_-_51065067 | 0.84 |
ENST00000595547.5
ENST00000335422.3 ENST00000595793.6 ENST00000596955.1 |
KLK13
|
kallikrein related peptidase 13 |
| chr3_+_172754457 | 0.83 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr5_+_136059151 | 0.81 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr10_+_17228215 | 0.80 |
ENST00000544301.7
|
VIM
|
vimentin |
| chr11_-_102798148 | 0.75 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1 |
| chr19_-_35513641 | 0.73 |
ENST00000339686.8
ENST00000447113.6 |
DMKN
|
dermokine |
| chr18_+_63775369 | 0.69 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr19_-_48511793 | 0.62 |
ENST00000600059.6
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr15_+_67166019 | 0.62 |
ENST00000537194.6
|
SMAD3
|
SMAD family member 3 |
| chr5_-_177496845 | 0.61 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr6_+_47698538 | 0.60 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr6_+_47698574 | 0.60 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr17_+_76385256 | 0.59 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr18_+_63775395 | 0.59 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr15_+_67125707 | 0.58 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr19_+_46601237 | 0.57 |
ENST00000597743.5
|
CALM3
|
calmodulin 3 |
| chr19_-_35501878 | 0.56 |
ENST00000593342.5
ENST00000601650.1 ENST00000408915.6 |
DMKN
|
dermokine |
| chr5_-_177496802 | 0.54 |
ENST00000506161.5
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr2_-_65366650 | 0.53 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr16_-_30113528 | 0.52 |
ENST00000406256.8
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr1_+_26280117 | 0.49 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamate rich protein like 3 |
| chr11_+_706595 | 0.47 |
ENST00000531348.5
ENST00000530636.5 |
EPS8L2
|
EPS8 like 2 |
| chr1_+_89364051 | 0.45 |
ENST00000370456.5
|
GBP6
|
guanylate binding protein family member 6 |
| chr1_+_22007450 | 0.45 |
ENST00000400271.2
|
CELA3A
|
chymotrypsin like elastase 3A |
| chr18_+_63476927 | 0.45 |
ENST00000489441.5
ENST00000382771.9 ENST00000424602.1 |
SERPINB5
|
serpin family B member 5 |
| chr4_+_83536097 | 0.44 |
ENST00000395226.6
ENST00000264409.5 |
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr19_-_38773432 | 0.44 |
ENST00000599035.1
ENST00000378626.5 |
LGALS7
|
galectin 7 |
| chr19_-_35490456 | 0.43 |
ENST00000338897.4
ENST00000484218.6 |
KRTDAP
|
keratinocyte differentiation associated protein |
| chr7_+_143381286 | 0.43 |
ENST00000449630.5
ENST00000322764.10 ENST00000457235.5 |
ZYX
|
zyxin |
| chr6_+_32153441 | 0.42 |
ENST00000414204.5
ENST00000361568.6 ENST00000395523.5 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr12_-_54419259 | 0.42 |
ENST00000293379.9
|
ITGA5
|
integrin subunit alpha 5 |
| chr7_+_134843884 | 0.41 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr11_-_64759967 | 0.41 |
ENST00000377432.7
ENST00000164139.4 |
PYGM
|
glycogen phosphorylase, muscle associated |
| chr17_-_19004727 | 0.40 |
ENST00000388995.11
|
FAM83G
|
family with sequence similarity 83 member G |
| chr14_+_94026314 | 0.40 |
ENST00000203664.10
ENST00000553723.1 |
OTUB2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr19_+_46601296 | 0.38 |
ENST00000598871.5
ENST00000291295.14 ENST00000594523.5 |
CALM3
|
calmodulin 3 |
| chr17_+_75721451 | 0.37 |
ENST00000200181.8
|
ITGB4
|
integrin subunit beta 4 |
| chr8_+_86098901 | 0.36 |
ENST00000285393.4
|
ATP6V0D2
|
ATPase H+ transporting V0 subunit d2 |
| chr20_-_653189 | 0.34 |
ENST00000381962.4
|
SRXN1
|
sulfiredoxin 1 |
| chr2_-_118847638 | 0.32 |
ENST00000295206.7
|
EN1
|
engrailed homeobox 1 |
| chr9_+_4985227 | 0.32 |
ENST00000381652.4
|
JAK2
|
Janus kinase 2 |
| chr6_-_43629222 | 0.32 |
ENST00000307126.10
|
GTPBP2
|
GTP binding protein 2 |
| chr17_-_41055211 | 0.31 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr8_-_124372686 | 0.31 |
ENST00000297632.8
|
TMEM65
|
transmembrane protein 65 |
| chr9_-_107489754 | 0.31 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr12_-_8612850 | 0.30 |
ENST00000229335.11
ENST00000537228.5 |
AICDA
|
activation induced cytidine deaminase |
| chr17_+_4950147 | 0.29 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr11_+_706196 | 0.29 |
ENST00000534755.5
ENST00000650127.1 |
EPS8L2
|
EPS8 like 2 |
| chr1_+_203026481 | 0.29 |
ENST00000367240.6
|
PPFIA4
|
PTPRF interacting protein alpha 4 |
| chr10_-_29634964 | 0.29 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr5_-_16916400 | 0.29 |
ENST00000513882.5
|
MYO10
|
myosin X |
| chr12_-_94616061 | 0.28 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr11_-_2903490 | 0.28 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chr1_+_156126160 | 0.27 |
ENST00000448611.6
ENST00000368297.5 |
LMNA
|
lamin A/C |
| chr5_+_14664653 | 0.27 |
ENST00000284274.5
|
OTULIN
|
OTU deubiquitinase with linear linkage specificity |
| chr3_-_125055987 | 0.26 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr19_-_43670153 | 0.26 |
ENST00000601723.5
ENST00000339082.7 ENST00000340093.8 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr10_+_84328625 | 0.26 |
ENST00000224756.12
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr11_+_706222 | 0.26 |
ENST00000318562.13
ENST00000533500.5 |
EPS8L2
|
EPS8 like 2 |
| chr8_-_142614469 | 0.25 |
ENST00000356613.4
|
ARC
|
activity regulated cytoskeleton associated protein |
| chr2_-_203535253 | 0.25 |
ENST00000457812.5
ENST00000319170.10 ENST00000630330.2 ENST00000308091.8 ENST00000453034.5 ENST00000420371.2 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr6_-_35921047 | 0.25 |
ENST00000361690.7
ENST00000512445.5 |
SRPK1
|
SRSF protein kinase 1 |
| chr11_+_706117 | 0.25 |
ENST00000533256.5
ENST00000614442.4 |
EPS8L2
|
EPS8 like 2 |
| chr4_-_88158605 | 0.25 |
ENST00000237612.8
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr11_-_102955705 | 0.24 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr17_-_8118489 | 0.24 |
ENST00000380149.6
ENST00000448843.7 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr4_-_79408198 | 0.24 |
ENST00000358842.5
|
GK2
|
glycerol kinase 2 |
| chr17_-_41786688 | 0.24 |
ENST00000310706.9
ENST00000393931.8 ENST00000424457.5 ENST00000591690.5 |
JUP
|
junction plakoglobin |
| chr7_+_2647703 | 0.23 |
ENST00000403167.5
|
TTYH3
|
tweety family member 3 |
| chr18_+_44700796 | 0.23 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr19_+_926001 | 0.22 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr6_-_35921079 | 0.22 |
ENST00000507909.1
ENST00000373825.7 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_-_35921128 | 0.22 |
ENST00000510290.5
ENST00000423325.6 |
SRPK1
|
SRSF protein kinase 1 |
| chrX_+_49171889 | 0.21 |
ENST00000376327.6
|
PLP2
|
proteolipid protein 2 |
| chr2_-_135876382 | 0.21 |
ENST00000264156.3
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr11_-_66336396 | 0.21 |
ENST00000627248.1
ENST00000311320.9 |
RIN1
|
Ras and Rab interactor 1 |
| chr8_-_142742398 | 0.20 |
ENST00000246515.2
|
SLURP1
|
secreted LY6/PLAUR domain containing 1 |
| chr17_-_35063648 | 0.20 |
ENST00000394597.7
|
RFFL
|
ring finger and FYVE like domain containing E3 ubiquitin protein ligase |
| chr11_-_11353241 | 0.20 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr8_-_67746348 | 0.20 |
ENST00000297770.10
|
CPA6
|
carboxypeptidase A6 |
| chr2_+_190343561 | 0.20 |
ENST00000322522.8
ENST00000392329.7 ENST00000430311.5 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr3_-_53844617 | 0.20 |
ENST00000481668.5
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr16_-_72172135 | 0.20 |
ENST00000537465.5
ENST00000237353.15 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr1_-_23799561 | 0.19 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr22_-_31662180 | 0.19 |
ENST00000435900.5
|
PISD
|
phosphatidylserine decarboxylase |
| chr5_-_79514127 | 0.19 |
ENST00000334082.11
|
HOMER1
|
homer scaffold protein 1 |
| chr6_+_125219804 | 0.19 |
ENST00000524679.1
|
TPD52L1
|
TPD52 like 1 |
| chr8_-_8893548 | 0.19 |
ENST00000276282.7
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr7_-_20217342 | 0.18 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr12_-_52406335 | 0.18 |
ENST00000257974.3
|
KRT82
|
keratin 82 |
| chr7_+_39623547 | 0.18 |
ENST00000005257.7
|
RALA
|
RAS like proto-oncogene A |
| chr17_-_29176752 | 0.18 |
ENST00000533112.5
|
MYO18A
|
myosin XVIIIA |
| chr11_-_95232514 | 0.18 |
ENST00000634898.1
ENST00000542176.1 ENST00000278499.6 |
SESN3
|
sestrin 3 |
| chr14_-_23155302 | 0.18 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr2_+_28392802 | 0.18 |
ENST00000379619.5
ENST00000264716.9 |
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr4_+_73740541 | 0.17 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr1_-_23799533 | 0.17 |
ENST00000429356.5
|
GALE
|
UDP-galactose-4-epimerase |
| chr8_-_109975757 | 0.17 |
ENST00000524391.6
|
KCNV1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr11_-_2171805 | 0.17 |
ENST00000381178.5
ENST00000381175.5 ENST00000333684.9 ENST00000352909.8 |
TH
|
tyrosine hydroxylase |
| chr10_+_84328581 | 0.17 |
ENST00000359979.8
ENST00000372088.8 |
CCSER2
|
coiled-coil serine rich protein 2 |
| chr6_+_159890964 | 0.16 |
ENST00000434562.2
ENST00000674077.2 |
ENSG00000236823.2
MAS1
|
novel transcript MAS1 proto-oncogene, G protein-coupled receptor |
| chr9_+_100429511 | 0.16 |
ENST00000613183.1
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr2_+_169069537 | 0.16 |
ENST00000428522.5
ENST00000450153.1 ENST00000674881.1 ENST00000421653.5 |
DHRS9
|
dehydrogenase/reductase 9 |
| chr17_-_81923532 | 0.16 |
ENST00000392366.7
|
MAFG
|
MAF bZIP transcription factor G |
| chr11_+_18599782 | 0.16 |
ENST00000634992.1
ENST00000635674.1 ENST00000511927.2 |
SPTY2D1OS
|
SPTY2D1 opposite strand |
| chr3_+_36380477 | 0.16 |
ENST00000457375.6
ENST00000273183.8 ENST00000434649.1 |
STAC
|
SH3 and cysteine rich domain |
| chr11_-_123885627 | 0.16 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chr6_-_656963 | 0.15 |
ENST00000380907.2
|
HUS1B
|
HUS1 checkpoint clamp component B |
| chr7_+_112423137 | 0.15 |
ENST00000005558.8
ENST00000621379.4 |
IFRD1
|
interferon related developmental regulator 1 |
| chr11_-_64246190 | 0.15 |
ENST00000392210.6
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr17_+_77319465 | 0.15 |
ENST00000329047.13
|
SEPTIN9
|
septin 9 |
| chr6_+_30717433 | 0.15 |
ENST00000681435.1
|
TUBB
|
tubulin beta class I |
| chr15_+_40239042 | 0.15 |
ENST00000558055.5
ENST00000455577.6 |
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr6_+_63571702 | 0.15 |
ENST00000672924.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr1_+_36155930 | 0.14 |
ENST00000316156.8
|
MAP7D1
|
MAP7 domain containing 1 |
| chr12_-_48957365 | 0.14 |
ENST00000398092.4
ENST00000539611.1 |
ENSG00000272822.2
ARF3
|
novel protein ADP ribosylation factor 3 |
| chr11_+_68903849 | 0.14 |
ENST00000675615.1
ENST00000255078.8 |
IGHMBP2
|
immunoglobulin mu DNA binding protein 2 |
| chr8_-_42768602 | 0.14 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr17_-_75405486 | 0.14 |
ENST00000392562.5
|
GRB2
|
growth factor receptor bound protein 2 |
| chr5_-_88731827 | 0.14 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr1_-_6602885 | 0.13 |
ENST00000377663.3
|
KLHL21
|
kelch like family member 21 |
| chr12_-_95996302 | 0.13 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr17_+_48892761 | 0.13 |
ENST00000355938.9
ENST00000393366.7 ENST00000503641.5 ENST00000514808.5 ENST00000506855.1 |
ATP5MC1
|
ATP synthase membrane subunit c locus 1 |
| chr15_+_40239420 | 0.13 |
ENST00000560346.5
|
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr9_+_98943705 | 0.13 |
ENST00000610452.1
|
COL15A1
|
collagen type XV alpha 1 chain |
| chr1_-_94541636 | 0.13 |
ENST00000370207.4
|
F3
|
coagulation factor III, tissue factor |
| chr20_+_43558968 | 0.13 |
ENST00000647834.1
ENST00000373100.7 ENST00000648083.1 ENST00000648530.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_+_36156096 | 0.13 |
ENST00000474796.2
ENST00000373150.8 ENST00000373151.6 |
MAP7D1
|
MAP7 domain containing 1 |
| chr19_+_38899680 | 0.13 |
ENST00000576510.5
ENST00000392079.7 |
NFKBIB
|
NFKB inhibitor beta |
| chr22_+_22720615 | 0.13 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr17_-_75405647 | 0.13 |
ENST00000316804.10
ENST00000316615.9 |
GRB2
|
growth factor receptor bound protein 2 |
| chr1_-_6602859 | 0.13 |
ENST00000377658.8
|
KLHL21
|
kelch like family member 21 |
| chr1_+_26863140 | 0.13 |
ENST00000339276.6
|
SFN
|
stratifin |
| chr5_+_66828762 | 0.13 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr16_+_89921851 | 0.12 |
ENST00000554444.5
ENST00000556565.5 |
TUBB3
|
tubulin beta 3 class III |
| chr22_-_38755990 | 0.12 |
ENST00000405018.5
ENST00000438058.5 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr9_-_74887720 | 0.12 |
ENST00000449912.6
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6 |
| chr1_-_94541746 | 0.12 |
ENST00000334047.12
|
F3
|
coagulation factor III, tissue factor |
| chrX_+_49171918 | 0.12 |
ENST00000376322.7
|
PLP2
|
proteolipid protein 2 |
| chr22_-_38755458 | 0.12 |
ENST00000405510.5
ENST00000433561.5 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr5_-_150086511 | 0.11 |
ENST00000675795.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr5_+_54455661 | 0.11 |
ENST00000302005.3
|
HSPB3
|
heat shock protein family B (small) member 3 |
| chr17_+_76738012 | 0.11 |
ENST00000590514.5
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr12_-_48957445 | 0.11 |
ENST00000541959.5
ENST00000447318.6 ENST00000256682.9 |
ARF3
|
ADP ribosylation factor 3 |
| chr17_-_75154534 | 0.10 |
ENST00000356033.8
|
JPT1
|
Jupiter microtubule associated homolog 1 |
| chr12_-_118190510 | 0.10 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr4_+_70721953 | 0.10 |
ENST00000381006.8
ENST00000226328.8 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_+_10456186 | 0.10 |
ENST00000528723.5
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr2_-_190062721 | 0.10 |
ENST00000260950.5
|
MSTN
|
myostatin |
| chr2_+_219572304 | 0.10 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha |
| chr17_-_29078857 | 0.10 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr15_+_75206398 | 0.10 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr14_-_75069478 | 0.10 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr8_-_42768781 | 0.10 |
ENST00000276410.7
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr3_-_98522514 | 0.10 |
ENST00000503004.5
ENST00000506575.1 ENST00000513452.5 ENST00000515620.5 |
CLDND1
|
claudin domain containing 1 |
| chr18_+_9885964 | 0.10 |
ENST00000357775.6
ENST00000306084.6 |
TXNDC2
|
thioredoxin domain containing 2 |
| chr17_+_7440738 | 0.10 |
ENST00000575398.5
ENST00000575082.5 |
FGF11
|
fibroblast growth factor 11 |
| chr6_-_38703066 | 0.09 |
ENST00000373365.5
|
GLO1
|
glyoxalase I |
| chr8_+_26293112 | 0.09 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr9_-_70414657 | 0.09 |
ENST00000377126.4
|
KLF9
|
Kruppel like factor 9 |
| chr9_-_74887914 | 0.09 |
ENST00000360774.6
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6 |
| chr3_-_31981228 | 0.09 |
ENST00000396556.7
ENST00000438237.6 |
OSBPL10
|
oxysterol binding protein like 10 |
| chr12_+_53299682 | 0.09 |
ENST00000267103.10
ENST00000548632.5 |
MYG1
|
MYG1 exonuclease |
| chr7_+_143222037 | 0.09 |
ENST00000408947.4
|
TAS2R40
|
taste 2 receptor member 40 |
| chr3_-_149576203 | 0.09 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_-_64245816 | 0.09 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr19_+_1407517 | 0.09 |
ENST00000336761.10
ENST00000233078.9 ENST00000592522.5 |
DAZAP1
|
DAZ associated protein 1 |
| chr20_-_17558811 | 0.09 |
ENST00000536626.7
ENST00000377868.6 |
BFSP1
|
beaded filament structural protein 1 |
| chr11_-_5516690 | 0.08 |
ENST00000380184.2
|
UBQLNL
|
ubiquilin like |
| chr11_+_128693887 | 0.08 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_168832005 | 0.08 |
ENST00000393726.7
ENST00000507735.6 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_-_47185840 | 0.08 |
ENST00000539589.5
ENST00000528462.5 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr3_-_169869833 | 0.08 |
ENST00000523069.1
ENST00000264676.9 ENST00000316428.10 |
LRRC31
|
leucine rich repeat containing 31 |
| chr2_-_96035974 | 0.08 |
ENST00000434632.5
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr7_-_42237187 | 0.08 |
ENST00000395925.8
|
GLI3
|
GLI family zinc finger 3 |
| chr8_-_27258414 | 0.07 |
ENST00000523048.5
|
STMN4
|
stathmin 4 |
| chr2_-_169031317 | 0.07 |
ENST00000650372.1
|
ABCB11
|
ATP binding cassette subfamily B member 11 |
| chr8_-_27258386 | 0.07 |
ENST00000350889.8
ENST00000519997.5 ENST00000519614.5 ENST00000522908.1 ENST00000265770.11 |
STMN4
|
stathmin 4 |
| chr20_+_36154630 | 0.07 |
ENST00000338074.7
ENST00000636016.2 ENST00000373945.5 |
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr7_-_32070396 | 0.07 |
ENST00000396191.6
|
PDE1C
|
phosphodiesterase 1C |
| chr3_+_148730100 | 0.07 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr9_-_35112379 | 0.07 |
ENST00000488109.6
|
FAM214B
|
family with sequence similarity 214 member B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.5 | 2.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.4 | 2.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 1.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 1.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 1.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.6 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 0.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.3 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 5.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 1.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.2 | GO:0052314 | phthalate metabolic process(GO:0018963) epinephrine biosynthetic process(GO:0042418) phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.8 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.7 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.3 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.4 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 1.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0060873 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 2.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 1.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 7.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 2.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.4 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.2 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 2.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.2 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.2 | GO:0034617 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |