Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
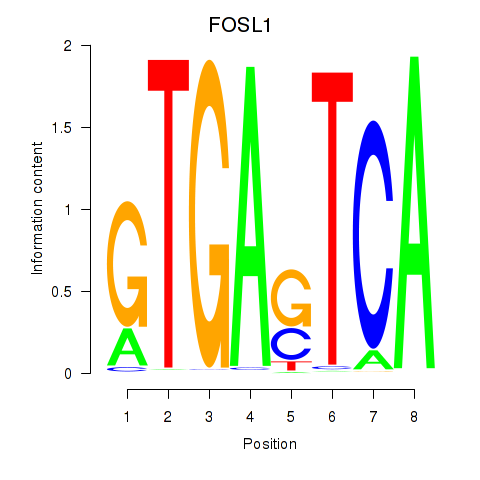
Results for FOSL1
Z-value: 2.15
Transcription factors associated with FOSL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL1
|
ENSG00000175592.9 | FOSL1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL1 | hg38_v1_chr11_-_65900413_65900424 | 0.88 | 1.8e-10 | Click! |
Activity profile of FOSL1 motif
Sorted Z-values of FOSL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_35528221 | 18.97 |
ENST00000588674.5
ENST00000452271.7 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_+_150508074 | 15.46 |
ENST00000369049.8
|
ECM1
|
extracellular matrix protein 1 |
| chr1_+_150508099 | 15.37 |
ENST00000346569.6
ENST00000369047.9 |
ECM1
|
extracellular matrix protein 1 |
| chr18_+_23873000 | 13.52 |
ENST00000269217.11
ENST00000587184.5 |
LAMA3
|
laminin subunit alpha 3 |
| chr1_+_183186238 | 12.29 |
ENST00000493293.5
ENST00000264144.5 |
LAMC2
|
laminin subunit gamma 2 |
| chr6_+_47698538 | 12.15 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr6_+_47698574 | 11.98 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr19_+_35154715 | 11.95 |
ENST00000392218.6
ENST00000543307.5 ENST00000392219.7 ENST00000541435.6 ENST00000590686.5 ENST00000342879.7 ENST00000588699.5 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr19_+_35154914 | 11.93 |
ENST00000423817.7
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr1_-_153057504 | 11.85 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A |
| chr12_+_53098846 | 11.31 |
ENST00000650247.1
ENST00000549628.1 |
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr10_+_17228215 | 10.66 |
ENST00000544301.7
|
VIM
|
vimentin |
| chr1_-_153041111 | 10.35 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr12_+_13196718 | 10.31 |
ENST00000431267.2
ENST00000542474.5 ENST00000544053.5 ENST00000256951.10 |
EMP1
|
epithelial membrane protein 1 |
| chr17_-_41612757 | 10.00 |
ENST00000301653.9
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr1_+_152908538 | 9.94 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr21_-_26843012 | 9.94 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 9.74 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr1_-_153549120 | 9.63 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_+_48088596 | 9.59 |
ENST00000416681.5
ENST00000331803.8 ENST00000432131.5 |
UPP1
|
uridine phosphorylase 1 |
| chr5_+_136059151 | 9.58 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr2_+_113117889 | 9.25 |
ENST00000361779.7
ENST00000259206.9 ENST00000354115.6 |
IL1RN
|
interleukin 1 receptor antagonist |
| chr11_-_65900375 | 9.17 |
ENST00000312562.7
|
FOSL1
|
FOS like 1, AP-1 transcription factor subunit |
| chr13_+_77535681 | 8.83 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535742 | 8.76 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr13_+_77535669 | 8.75 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr9_+_35673917 | 8.01 |
ENST00000617161.1
ENST00000378357.9 |
CA9
|
carbonic anhydrase 9 |
| chr11_-_65900413 | 7.86 |
ENST00000448083.6
ENST00000531493.5 ENST00000532401.1 |
FOSL1
|
FOS like 1, AP-1 transcription factor subunit |
| chr17_+_76385256 | 7.66 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr1_-_153549238 | 7.65 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_-_205449924 | 7.61 |
ENST00000367154.5
|
LEMD1
|
LEM domain containing 1 |
| chr16_+_50266530 | 7.56 |
ENST00000566433.6
ENST00000394697.7 ENST00000673801.1 |
ADCY7
|
adenylate cyclase 7 |
| chr12_-_119804472 | 7.13 |
ENST00000678087.1
ENST00000677993.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr12_-_119804298 | 7.13 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr11_-_62556230 | 6.21 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr20_+_35172046 | 6.15 |
ENST00000216968.5
|
PROCR
|
protein C receptor |
| chr6_+_73695779 | 6.07 |
ENST00000422508.6
ENST00000437994.6 |
CD109
|
CD109 molecule |
| chr11_-_102798148 | 6.07 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1 |
| chr11_+_35176696 | 5.97 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr6_+_73696145 | 5.96 |
ENST00000287097.6
|
CD109
|
CD109 molecule |
| chr1_-_204151884 | 5.74 |
ENST00000367201.7
|
ETNK2
|
ethanolamine kinase 2 |
| chr7_-_108003122 | 5.54 |
ENST00000393559.2
ENST00000222399.11 ENST00000676777.1 ENST00000439976.6 ENST00000393560.5 ENST00000677793.1 ENST00000679244.1 |
LAMB1
|
laminin subunit beta 1 |
| chr3_-_48595267 | 5.52 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr4_-_39032343 | 5.43 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr11_-_6320494 | 5.42 |
ENST00000303927.4
ENST00000530979.1 |
CAVIN3
|
caveolae associated protein 3 |
| chr17_+_4950147 | 5.41 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr9_-_120877026 | 5.36 |
ENST00000436309.5
|
PHF19
|
PHD finger protein 19 |
| chr19_-_35501878 | 5.35 |
ENST00000593342.5
ENST00000601650.1 ENST00000408915.6 |
DMKN
|
dermokine |
| chr1_-_204152010 | 5.32 |
ENST00000367202.9
|
ETNK2
|
ethanolamine kinase 2 |
| chr19_-_43781249 | 5.21 |
ENST00000615047.4
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr11_+_35176575 | 5.18 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_35176611 | 5.05 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_35176639 | 5.03 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr18_+_63775395 | 4.99 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr1_-_28193873 | 4.96 |
ENST00000305392.3
ENST00000539896.1 |
PTAFR
|
platelet activating factor receptor |
| chr18_+_63775369 | 4.92 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr19_-_35513641 | 4.89 |
ENST00000339686.8
ENST00000447113.6 |
DMKN
|
dermokine |
| chr7_-_23347704 | 4.86 |
ENST00000619562.4
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr12_-_54419259 | 4.86 |
ENST00000293379.9
|
ITGA5
|
integrin subunit alpha 5 |
| chr19_-_43780957 | 4.81 |
ENST00000648319.1
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr19_-_51034840 | 4.78 |
ENST00000529888.5
|
KLK12
|
kallikrein related peptidase 12 |
| chr19_-_51034892 | 4.69 |
ENST00000319590.8
ENST00000250351.4 |
KLK12
|
kallikrein related peptidase 12 |
| chr6_+_106098933 | 4.66 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr19_-_51020154 | 4.62 |
ENST00000391805.5
ENST00000599077.1 |
KLK10
|
kallikrein related peptidase 10 |
| chr12_-_94616061 | 4.62 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr11_-_66336396 | 4.55 |
ENST00000627248.1
ENST00000311320.9 |
RIN1
|
Ras and Rab interactor 1 |
| chr1_-_150235972 | 4.52 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr1_-_154970735 | 4.45 |
ENST00000368445.9
ENST00000448116.7 ENST00000368449.8 |
SHC1
|
SHC adaptor protein 1 |
| chr19_-_51034993 | 4.42 |
ENST00000684732.1
|
KLK12
|
kallikrein related peptidase 12 |
| chr5_+_136058849 | 4.39 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr7_+_143381286 | 4.23 |
ENST00000449630.5
ENST00000322764.10 ENST00000457235.5 |
ZYX
|
zyxin |
| chr1_+_153031195 | 4.14 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B |
| chr4_-_39032922 | 4.02 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr11_+_706595 | 3.99 |
ENST00000531348.5
ENST00000530636.5 |
EPS8L2
|
EPS8 like 2 |
| chr10_-_88851809 | 3.92 |
ENST00000371930.5
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr19_-_51020019 | 3.91 |
ENST00000309958.7
|
KLK10
|
kallikrein related peptidase 10 |
| chr12_-_95116967 | 3.91 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr10_-_75109085 | 3.81 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr1_-_162412117 | 3.80 |
ENST00000367929.3
|
SH2D1B
|
SH2 domain containing 1B |
| chr9_+_137225166 | 3.77 |
ENST00000650725.2
|
CYSRT1
|
cysteine rich tail 1 |
| chr2_+_219627394 | 3.76 |
ENST00000373760.6
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr4_+_83535914 | 3.70 |
ENST00000611707.4
|
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr11_-_102955705 | 3.64 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr4_+_83536097 | 3.61 |
ENST00000395226.6
ENST00000264409.5 |
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr2_+_219627650 | 3.48 |
ENST00000317151.7
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr9_-_35112379 | 3.46 |
ENST00000488109.6
|
FAM214B
|
family with sequence similarity 214 member B |
| chr1_-_94541636 | 3.44 |
ENST00000370207.4
|
F3
|
coagulation factor III, tissue factor |
| chr2_+_219627565 | 3.39 |
ENST00000273063.10
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr3_-_98522514 | 3.39 |
ENST00000503004.5
ENST00000506575.1 ENST00000513452.5 ENST00000515620.5 |
CLDND1
|
claudin domain containing 1 |
| chr2_+_219627622 | 3.34 |
ENST00000358055.8
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr12_-_53232182 | 3.33 |
ENST00000425354.7
ENST00000546717.1 ENST00000394426.5 |
RARG
|
retinoic acid receptor gamma |
| chr1_+_24319342 | 3.31 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr1_+_24319511 | 3.29 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr17_-_41624541 | 3.20 |
ENST00000540235.5
ENST00000311208.13 |
KRT17
|
keratin 17 |
| chr1_-_94541746 | 3.20 |
ENST00000334047.12
|
F3
|
coagulation factor III, tissue factor |
| chr2_+_190927649 | 3.20 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr19_+_926001 | 3.18 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr16_-_28506826 | 3.17 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr16_+_58500047 | 3.16 |
ENST00000566192.5
ENST00000565088.5 ENST00000568640.5 |
NDRG4
|
NDRG family member 4 |
| chr16_-_30113528 | 3.05 |
ENST00000406256.8
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr18_+_63887698 | 3.04 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr5_+_150497772 | 3.01 |
ENST00000523767.5
|
NDST1
|
N-deacetylase and N-sulfotransferase 1 |
| chr6_+_41637005 | 2.99 |
ENST00000419164.6
ENST00000373051.6 |
MDFI
|
MyoD family inhibitor |
| chr1_-_150235943 | 2.98 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr1_-_151992571 | 2.93 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr1_-_150235995 | 2.93 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr15_-_70096604 | 2.90 |
ENST00000559048.5
ENST00000560939.5 ENST00000440567.7 ENST00000557907.5 ENST00000558379.5 ENST00000559929.5 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr17_+_75721327 | 2.90 |
ENST00000579662.5
|
ITGB4
|
integrin subunit beta 4 |
| chr15_-_70096260 | 2.89 |
ENST00000558201.5
|
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr2_-_65366650 | 2.84 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr3_-_151316795 | 2.79 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr17_-_9791586 | 2.78 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chr11_+_706196 | 2.78 |
ENST00000534755.5
ENST00000650127.1 |
EPS8L2
|
EPS8 like 2 |
| chr11_+_394196 | 2.77 |
ENST00000331563.7
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr16_+_58500135 | 2.75 |
ENST00000563978.5
ENST00000569923.5 ENST00000356752.8 ENST00000563799.5 ENST00000562999.5 ENST00000570248.5 ENST00000562731.5 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr2_+_33134620 | 2.72 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_+_75721471 | 2.69 |
ENST00000450894.7
|
ITGB4
|
integrin subunit beta 4 |
| chr16_+_83953232 | 2.69 |
ENST00000565123.5
ENST00000393306.6 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr5_-_140633690 | 2.65 |
ENST00000512545.1
ENST00000401743.6 |
CD14
|
CD14 molecule |
| chr9_+_4985227 | 2.64 |
ENST00000381652.4
|
JAK2
|
Janus kinase 2 |
| chr17_-_47851155 | 2.63 |
ENST00000536300.2
|
SP6
|
Sp6 transcription factor |
| chr11_-_2903490 | 2.61 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chrX_+_100644183 | 2.61 |
ENST00000640889.1
ENST00000373004.5 |
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr2_+_28392802 | 2.60 |
ENST00000379619.5
ENST00000264716.9 |
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr4_-_993430 | 2.59 |
ENST00000361661.6
ENST00000622731.4 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr11_+_706222 | 2.55 |
ENST00000318562.13
ENST00000533500.5 |
EPS8L2
|
EPS8 like 2 |
| chr6_+_106086316 | 2.53 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr6_+_32153441 | 2.52 |
ENST00000414204.5
ENST00000361568.6 ENST00000395523.5 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr19_+_38789198 | 2.50 |
ENST00000314980.5
|
LGALS7B
|
galectin 7B |
| chr1_+_155033824 | 2.49 |
ENST00000295542.6
ENST00000423025.6 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chrX_+_49171889 | 2.47 |
ENST00000376327.6
|
PLP2
|
proteolipid protein 2 |
| chr2_+_33134579 | 2.46 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr11_+_706117 | 2.45 |
ENST00000533256.5
ENST00000614442.4 |
EPS8L2
|
EPS8 like 2 |
| chr3_-_149377637 | 2.43 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr8_-_124372686 | 2.41 |
ENST00000297632.8
|
TMEM65
|
transmembrane protein 65 |
| chr3_+_191329020 | 2.41 |
ENST00000392456.4
|
CCDC50
|
coiled-coil domain containing 50 |
| chrX_-_71068384 | 2.39 |
ENST00000276105.3
ENST00000622259.4 |
SNX12
|
sorting nexin 12 |
| chr7_+_128455849 | 2.36 |
ENST00000435296.2
ENST00000257696.5 |
HILPDA
|
hypoxia inducible lipid droplet associated |
| chr9_+_121567057 | 2.32 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr14_+_64504574 | 2.31 |
ENST00000358738.3
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr9_-_35111573 | 2.26 |
ENST00000378561.5
ENST00000603301.5 |
FAM214B
|
family with sequence similarity 214 member B |
| chr11_-_82997477 | 2.24 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr12_-_47758828 | 2.23 |
ENST00000389212.7
ENST00000449771.7 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr10_-_75109172 | 2.21 |
ENST00000372700.7
ENST00000473072.2 ENST00000491677.6 ENST00000372702.7 |
DUSP13
|
dual specificity phosphatase 13 |
| chr7_+_134843884 | 2.20 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr17_-_45425620 | 2.19 |
ENST00000376922.6
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr12_-_57520480 | 2.19 |
ENST00000642841.1
ENST00000547303.5 ENST00000552740.5 ENST00000547526.1 ENST00000346473.8 ENST00000551116.5 |
ENSG00000285133.1
DDIT3
|
novel protein DNA damage inducible transcript 3 |
| chr8_-_42768602 | 2.19 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr14_+_96256194 | 2.18 |
ENST00000216629.11
ENST00000553356.1 |
BDKRB1
|
bradykinin receptor B1 |
| chr8_+_125430333 | 2.16 |
ENST00000311922.4
|
TRIB1
|
tribbles pseudokinase 1 |
| chr6_-_43629222 | 2.16 |
ENST00000307126.10
|
GTPBP2
|
GTP binding protein 2 |
| chr9_-_35111423 | 2.15 |
ENST00000378557.1
|
FAM214B
|
family with sequence similarity 214 member B |
| chr3_+_57060658 | 2.15 |
ENST00000334325.2
|
SPATA12
|
spermatogenesis associated 12 |
| chr12_+_6914571 | 2.14 |
ENST00000229277.6
ENST00000538763.5 ENST00000545045.6 |
ENO2
|
enolase 2 |
| chr8_-_42768781 | 2.13 |
ENST00000276410.7
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr14_+_94026314 | 2.11 |
ENST00000203664.10
ENST00000553723.1 |
OTUB2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr15_+_40239042 | 2.08 |
ENST00000558055.5
ENST00000455577.6 |
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr1_+_61952283 | 2.08 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr18_+_44700796 | 2.06 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr15_-_55917080 | 2.04 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr10_+_30434021 | 2.03 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr16_-_2858154 | 2.02 |
ENST00000571228.1
ENST00000161006.8 |
PRSS22
|
serine protease 22 |
| chr1_-_155978144 | 2.00 |
ENST00000313695.11
ENST00000497907.5 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr17_-_41586887 | 2.00 |
ENST00000167586.7
|
KRT14
|
keratin 14 |
| chr11_+_73289403 | 1.98 |
ENST00000535931.2
ENST00000544437.6 |
P2RY6
|
pyrimidinergic receptor P2Y6 |
| chr17_-_28717741 | 1.98 |
ENST00000395243.7
|
RAB34
|
RAB34, member RAS oncogene family |
| chr9_-_107489754 | 1.98 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr6_-_35921047 | 1.98 |
ENST00000361690.7
ENST00000512445.5 |
SRPK1
|
SRSF protein kinase 1 |
| chr9_-_127578989 | 1.96 |
ENST00000373314.7
|
NIBAN2
|
niban apoptosis regulator 2 |
| chr15_-_55917129 | 1.94 |
ENST00000338963.6
ENST00000508342.5 |
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr15_+_88638947 | 1.94 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr18_+_54828406 | 1.94 |
ENST00000262094.10
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr17_-_35063648 | 1.92 |
ENST00000394597.7
|
RFFL
|
ring finger and FYVE like domain containing E3 ubiquitin protein ligase |
| chr12_-_76084612 | 1.91 |
ENST00000535020.6
ENST00000552342.5 |
NAP1L1
|
nucleosome assembly protein 1 like 1 |
| chr4_+_73740541 | 1.90 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr1_+_156126160 | 1.90 |
ENST00000448611.6
ENST00000368297.5 |
LMNA
|
lamin A/C |
| chr15_+_40239420 | 1.90 |
ENST00000560346.5
|
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr10_+_30434176 | 1.90 |
ENST00000263056.6
ENST00000375322.2 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr3_-_37174578 | 1.88 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr17_-_28718405 | 1.87 |
ENST00000430132.6
ENST00000301043.10 ENST00000412625.5 |
RAB34
|
RAB34, member RAS oncogene family |
| chr15_-_70702273 | 1.87 |
ENST00000558758.5
ENST00000379983.6 ENST00000560441.5 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr1_+_26280117 | 1.86 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamate rich protein like 3 |
| chr4_-_79408198 | 1.82 |
ENST00000358842.5
|
GK2
|
glycerol kinase 2 |
| chr6_-_83431038 | 1.79 |
ENST00000369705.4
|
ME1
|
malic enzyme 1 |
| chr2_+_201451711 | 1.79 |
ENST00000194530.8
ENST00000392249.6 |
STRADB
|
STE20 related adaptor beta |
| chr2_-_164841410 | 1.79 |
ENST00000342193.8
ENST00000375458.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr1_-_150236064 | 1.78 |
ENST00000532744.2
ENST00000369114.9 ENST00000369115.3 ENST00000583931.6 |
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr6_-_35921079 | 1.76 |
ENST00000507909.1
ENST00000373825.7 |
SRPK1
|
SRSF protein kinase 1 |
| chr17_-_19004727 | 1.75 |
ENST00000388995.11
|
FAM83G
|
family with sequence similarity 83 member G |
| chr11_+_35618450 | 1.75 |
ENST00000317811.6
|
FJX1
|
four-jointed box kinase 1 |
| chr19_-_40257045 | 1.73 |
ENST00000578615.6
|
AKT2
|
AKT serine/threonine kinase 2 |
| chr6_-_35921128 | 1.73 |
ENST00000510290.5
ENST00000423325.6 |
SRPK1
|
SRSF protein kinase 1 |
| chr7_-_42237187 | 1.73 |
ENST00000395925.8
|
GLI3
|
GLI family zinc finger 3 |
| chr8_-_67746348 | 1.72 |
ENST00000297770.10
|
CPA6
|
carboxypeptidase A6 |
| chr20_-_17558811 | 1.70 |
ENST00000536626.7
ENST00000377868.6 |
BFSP1
|
beaded filament structural protein 1 |
| chr9_-_124415421 | 1.67 |
ENST00000259457.8
ENST00000441097.1 |
PSMB7
|
proteasome 20S subunit beta 7 |
| chr19_+_45469841 | 1.66 |
ENST00000592811.5
ENST00000586615.5 |
FOSB
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr11_-_47185840 | 1.65 |
ENST00000539589.5
ENST00000528462.5 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr20_+_46008900 | 1.60 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr3_-_195876635 | 1.58 |
ENST00000672669.1
ENST00000672886.1 ENST00000672098.1 ENST00000671767.1 ENST00000672548.1 |
TNK2
|
tyrosine kinase non receptor 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 23.9 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 3.2 | 9.6 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 2.9 | 17.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 2.6 | 10.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 2.6 | 7.7 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 2.5 | 9.9 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 2.4 | 12.0 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 2.4 | 7.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 2.0 | 9.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 2.0 | 2.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 1.8 | 5.4 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 1.7 | 6.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.6 | 21.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.3 | 4.0 | GO:0010768 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 1.2 | 34.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.2 | 3.5 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 1.1 | 3.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 1.0 | 30.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 1.0 | 5.0 | GO:1902943 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 1.0 | 3.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.9 | 7.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.9 | 2.6 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.9 | 15.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.9 | 3.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 2.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.7 | 11.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.7 | 2.9 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.7 | 2.2 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.7 | 10.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.7 | 10.7 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.6 | 5.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.6 | 11.6 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.6 | 1.7 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.6 | 1.7 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.6 | 1.7 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.5 | 5.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.5 | 3.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.5 | 2.6 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.5 | 1.5 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.5 | 7.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.5 | 4.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 2.5 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.5 | 2.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.5 | 1.5 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.4 | 5.8 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.4 | 11.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 24.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 2.0 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 1.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.4 | 2.3 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.4 | 1.9 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 10.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 7.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 3.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 1.8 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) |
| 0.4 | 6.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 2.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.3 | 1.3 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.3 | 21.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 5.9 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 1.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 3.3 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 2.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 1.2 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 1.8 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.3 | 2.1 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 11.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 1.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 2.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 0.8 | GO:0045553 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.3 | 4.9 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.3 | 2.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 1.8 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 2.2 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.2 | 13.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 5.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 4.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 14.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 3.0 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 2.7 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.2 | 2.9 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 0.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 3.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 3.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 2.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 11.2 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 0.6 | GO:0042946 | glucoside transport(GO:0042946) |
| 0.2 | 2.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.7 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.2 | 1.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 3.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 1.9 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 0.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.8 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 1.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 4.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.5 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 2.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 14.4 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 9.3 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 20.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 2.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.5 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 1.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 1.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 4.4 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.9 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.6 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 1.0 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 1.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 7.9 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.3 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.3 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 14.0 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 2.6 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 0.9 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.8 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 3.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 5.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 1.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 3.0 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 1.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.0 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) |
| 0.1 | 0.9 | GO:1903543 | regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) |
| 0.1 | 0.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.3 | GO:1990182 | exosomal secretion(GO:1990182) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 2.2 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of cellular pH reduction(GO:0032849) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 3.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.2 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.1 | 1.5 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 0.7 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 2.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 1.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.1 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 1.0 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 2.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0060411 | cardiac septum morphogenesis(GO:0060411) |
| 0.0 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 2.7 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.0 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 2.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 1.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.4 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 1.1 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 2.3 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.6 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 1.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 1.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 2.4 | GO:0003231 | cardiac ventricle development(GO:0003231) |
| 0.0 | 1.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.8 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.2 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.8 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 1.7 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 1.1 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 12.1 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 32.6 | GO:0043256 | laminin complex(GO:0043256) |
| 1.9 | 21.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.6 | 4.9 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.5 | 30.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.2 | 67.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 1.1 | 4.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.9 | 11.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.9 | 5.5 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.8 | 7.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.7 | 2.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.7 | 13.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.6 | 2.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.4 | 7.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 5.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 2.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 13.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 0.9 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.3 | 2.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 3.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 5.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 7.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 30.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 1.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 6.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 0.8 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.2 | 5.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 30.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 3.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 24.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.2 | 7.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 4.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.7 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 14.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 12.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 3.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 9.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 4.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 5.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 6.2 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 11.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 4.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 29.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 3.2 | 9.6 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 3.1 | 9.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 2.1 | 12.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.6 | 11.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 1.2 | 7.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.2 | 3.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.1 | 4.5 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 1.1 | 7.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 1.0 | 5.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.0 | 10.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 11.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.8 | 5.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 7.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.8 | 3.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.7 | 2.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 5.6 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.6 | 12.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.6 | 7.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.5 | 3.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 21.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 2.5 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.5 | 4.0 | GO:0050816 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.5 | 3.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 7.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 2.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.4 | 2.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.4 | 1.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.4 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 2.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 1.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.4 | 12.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.4 | 1.8 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 2.0 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.3 | 1.3 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.3 | 12.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 5.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 2.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 3.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 18.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 7.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 0.7 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.2 | 1.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 6.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 2.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 16.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 0.6 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 0.2 | 4.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 2.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 0.7 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.2 | 1.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 7.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 3.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 2.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 4.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 2.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 0.5 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 4.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 1.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 2.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 11.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 23.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 12.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 15.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 2.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 3.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 27.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.2 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 2.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 2.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 4.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 4.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 5.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.7 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 5.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 1.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.7 | GO:1990239 | estrogen receptor activity(GO:0030284) steroid hormone binding(GO:1990239) |
| 0.1 | 1.5 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 13.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 5.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 2.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 6.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 16.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 18.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 38.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 22.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 35.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 20.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 7.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 9.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 2.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 3.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 7.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 8.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 8.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 43.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 11.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 3.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 9.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 6.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 12.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 35.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 5.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 8.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 11.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 21.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.7 | 17.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 9.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 11.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.5 | 7.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 16.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 9.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 5.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 2.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 31.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 4.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 4.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 12.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 2.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 11.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 4.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 7.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 6.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 4.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.7 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 2.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 12.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 5.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 6.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 12.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 11.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 5.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 6.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 3.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 4.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |