|
chr20_+_33235987
Show fit
|
4.67 |
ENST00000375422.6
ENST00000375413.8
ENST00000354297.9
|
BPIFA1
|
BPI fold containing family A member 1
|
|
chr4_+_164754045
Show fit
|
4.24 |
ENST00000515485.5
|
SMIM31
|
small integral membrane protein 31
|
|
chr4_+_164754116
Show fit
|
3.32 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31
|
|
chr10_-_46046264
Show fit
|
2.39 |
ENST00000581478.5
ENST00000582163.3
|
MSMB
|
microseminoprotein beta
|
|
chr12_-_71157992
Show fit
|
1.76 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8
|
|
chr12_-_71157872
Show fit
|
1.73 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8
|
|
chr2_+_70935919
Show fit
|
1.65 |
ENST00000412314.5
|
ATP6V1B1
|
ATPase H+ transporting V1 subunit B1
|
|
chr2_+_70935864
Show fit
|
1.65 |
ENST00000234396.10
ENST00000454446.6
|
ATP6V1B1
|
ATPase H+ transporting V1 subunit B1
|
|
chr9_-_86947496
Show fit
|
1.56 |
ENST00000298743.9
|
GAS1
|
growth arrest specific 1
|
|
chr7_+_80602150
Show fit
|
1.54 |
ENST00000309881.11
|
CD36
|
CD36 molecule
|
|
chr7_+_80602200
Show fit
|
1.53 |
ENST00000534394.5
|
CD36
|
CD36 molecule
|
|
chr18_+_3449413
Show fit
|
1.52 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr6_-_46954922
Show fit
|
1.51 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5
|
|
chr13_-_85799400
Show fit
|
1.38 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6
|
|
chr11_-_124752247
Show fit
|
1.38 |
ENST00000326621.10
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr1_+_87331668
Show fit
|
1.31 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4
|
|
chr11_-_124752187
Show fit
|
1.28 |
ENST00000403470.1
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr3_+_186996444
Show fit
|
1.21 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1
|
|
chr9_-_14722725
Show fit
|
1.20 |
ENST00000380911.4
|
CER1
|
cerberus 1, DAN family BMP antagonist
|
|
chrX_-_15600953
Show fit
|
1.17 |
ENST00000679212.1
ENST00000679278.1
ENST00000678046.1
ENST00000252519.8
|
ACE2
|
angiotensin I converting enzyme 2
|
|
chr20_+_33168148
Show fit
|
1.12 |
ENST00000354932.6
|
BPIFA2
|
BPI fold containing family A member 2
|
|
chr1_-_206921987
Show fit
|
1.07 |
ENST00000530505.1
ENST00000442471.4
|
FCMR
|
Fc fragment of IgM receptor
|
|
chr20_-_57266342
Show fit
|
1.01 |
ENST00000395864.7
|
BMP7
|
bone morphogenetic protein 7
|
|
chr11_-_108593738
Show fit
|
0.96 |
ENST00000525344.5
ENST00000265843.9
|
EXPH5
|
exophilin 5
|
|
chr1_-_206921867
Show fit
|
0.89 |
ENST00000628511.2
ENST00000367091.8
|
FCMR
|
Fc fragment of IgM receptor
|
|
chr15_+_60004305
Show fit
|
0.80 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1
|
|
chrX_-_15601077
Show fit
|
0.76 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2
|
|
chr17_-_47957824
Show fit
|
0.76 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like
|
|
chr1_+_61081728
Show fit
|
0.76 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A
|
|
chr14_-_21025490
Show fit
|
0.75 |
ENST00000553442.5
ENST00000555869.5
ENST00000556457.5
ENST00000397844.6
ENST00000554415.5
ENST00000556974.5
ENST00000554419.5
ENST00000298687.9
ENST00000397858.5
ENST00000360463.7
ENST00000350792.7
ENST00000397847.6
|
NDRG2
|
NDRG family member 2
|
|
chr3_-_149221811
Show fit
|
0.71 |
ENST00000455472.3
ENST00000264613.11
|
CP
|
ceruloplasmin
|
|
chr12_-_91178520
Show fit
|
0.70 |
ENST00000425043.5
ENST00000420120.6
ENST00000441303.6
ENST00000456569.2
|
DCN
|
decorin
|
|
chr2_+_168901290
Show fit
|
0.66 |
ENST00000429379.2
ENST00000375363.8
ENST00000421979.1
|
G6PC2
|
glucose-6-phosphatase catalytic subunit 2
|
|
chr19_-_48615050
Show fit
|
0.64 |
ENST00000263266.4
|
FAM83E
|
family with sequence similarity 83 member E
|
|
chr20_-_57266606
Show fit
|
0.64 |
ENST00000450594.6
ENST00000395863.8
|
BMP7
|
bone morphogenetic protein 7
|
|
chr19_+_42302098
Show fit
|
0.64 |
ENST00000598490.1
ENST00000341747.8
|
PRR19
|
proline rich 19
|
|
chr20_-_54070520
Show fit
|
0.63 |
ENST00000371435.6
ENST00000395961.7
|
BCAS1
|
brain enriched myelin associated protein 1
|
|
chr3_-_193554885
Show fit
|
0.63 |
ENST00000342695.9
|
ATP13A4
|
ATPase 13A4
|
|
chr10_+_122560639
Show fit
|
0.62 |
ENST00000344338.7
ENST00000330163.8
ENST00000652446.2
ENST00000666315.1
ENST00000368955.7
ENST00000368909.7
ENST00000368956.6
ENST00000619379.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr5_+_140806929
Show fit
|
0.62 |
ENST00000378125.4
ENST00000618834.1
ENST00000530339.2
ENST00000512229.6
ENST00000672575.1
|
PCDHA4
|
protocadherin alpha 4
|
|
chr1_+_61082398
Show fit
|
0.61 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A
|
|
chr17_+_9021501
Show fit
|
0.61 |
ENST00000173229.7
|
NTN1
|
netrin 1
|
|
chr5_+_102865805
Show fit
|
0.60 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr11_+_66258467
Show fit
|
0.60 |
ENST00000394066.6
|
KLC2
|
kinesin light chain 2
|
|
chr10_+_122560679
Show fit
|
0.58 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr7_+_114414809
Show fit
|
0.55 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2
|
|
chr20_+_45306834
Show fit
|
0.54 |
ENST00000343694.8
ENST00000372741.7
ENST00000372743.5
|
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region like
|
|
chr13_-_48413105
Show fit
|
0.54 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr3_-_193554952
Show fit
|
0.52 |
ENST00000392443.7
|
ATP13A4
|
ATPase 13A4
|
|
chr1_+_61082553
Show fit
|
0.51 |
ENST00000403491.8
ENST00000371187.7
|
NFIA
|
nuclear factor I A
|
|
chr10_+_122560751
Show fit
|
0.51 |
ENST00000338354.10
ENST00000664692.1
ENST00000653442.1
ENST00000664974.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr14_+_104801082
Show fit
|
0.51 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr1_-_201127184
Show fit
|
0.49 |
ENST00000449188.3
|
ASCL5
|
achaete-scute family bHLH transcription factor 5
|
|
chr4_+_186227501
Show fit
|
0.48 |
ENST00000446598.6
ENST00000264690.11
ENST00000513864.2
|
KLKB1
|
kallikrein B1
|
|
chr9_+_102995308
Show fit
|
0.47 |
ENST00000612124.4
ENST00000374798.8
ENST00000487798.5
|
CYLC2
|
cylicin 2
|
|
chr12_+_10212867
Show fit
|
0.47 |
ENST00000545047.5
ENST00000266458.10
ENST00000629504.1
ENST00000543602.5
ENST00000545887.1
|
GABARAPL1
|
GABA type A receptor associated protein like 1
|
|
chr14_-_69797232
Show fit
|
0.47 |
ENST00000216540.5
|
SLC10A1
|
solute carrier family 10 member 1
|
|
chr8_+_94895813
Show fit
|
0.46 |
ENST00000396113.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6
|
|
chr16_-_4846196
Show fit
|
0.45 |
ENST00000589389.5
|
GLYR1
|
glyoxylate reductase 1 homolog
|
|
chrX_+_16946862
Show fit
|
0.45 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr21_-_42315336
Show fit
|
0.45 |
ENST00000398431.2
ENST00000518498.3
|
TFF3
|
trefoil factor 3
|
|
chr17_+_9825906
Show fit
|
0.44 |
ENST00000262441.10
|
GLP2R
|
glucagon like peptide 2 receptor
|
|
chr3_-_193554799
Show fit
|
0.43 |
ENST00000295548.3
|
ATP13A4
|
ATPase 13A4
|
|
chr11_+_35662739
Show fit
|
0.42 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44
|
|
chr8_+_94895763
Show fit
|
0.40 |
ENST00000523378.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6
|
|
chr9_-_20622479
Show fit
|
0.40 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit
|
|
chr3_+_69936583
Show fit
|
0.39 |
ENST00000314557.10
ENST00000394351.9
|
MITF
|
melanocyte inducing transcription factor
|
|
chr2_-_171160833
Show fit
|
0.39 |
ENST00000360843.7
ENST00000431350.7
|
TLK1
|
tousled like kinase 1
|
|
chr10_+_122163672
Show fit
|
0.39 |
ENST00000369004.7
ENST00000260733.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr3_+_148827800
Show fit
|
0.39 |
ENST00000282957.9
ENST00000468341.1
|
CPB1
|
carboxypeptidase B1
|
|
chr6_-_42048648
Show fit
|
0.39 |
ENST00000502771.1
ENST00000508143.5
ENST00000514588.1
ENST00000510503.5
|
CCND3
|
cyclin D3
|
|
chr15_+_96325935
Show fit
|
0.38 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2
|
|
chr4_+_85475131
Show fit
|
0.38 |
ENST00000395184.6
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr22_-_21227637
Show fit
|
0.38 |
ENST00000401924.5
|
GGT2
|
gamma-glutamyltransferase 2
|
|
chr10_+_122163590
Show fit
|
0.38 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr4_+_169660062
Show fit
|
0.37 |
ENST00000507875.5
ENST00000613795.4
|
CLCN3
|
chloride voltage-gated channel 3
|
|
chr8_-_132760548
Show fit
|
0.37 |
ENST00000519187.5
ENST00000523829.5
ENST00000677595.1
ENST00000356838.7
ENST00000377901.8
ENST00000519304.1
|
TMEM71
|
transmembrane protein 71
|
|
chr4_+_74308463
Show fit
|
0.36 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen
|
|
chr2_+_203936755
Show fit
|
0.35 |
ENST00000316386.11
ENST00000435193.1
|
ICOS
|
inducible T cell costimulator
|
|
chr1_+_222928415
Show fit
|
0.34 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1
|
|
chr2_-_213151590
Show fit
|
0.33 |
ENST00000374319.8
ENST00000457361.5
ENST00000451136.6
ENST00000434687.6
|
IKZF2
|
IKAROS family zinc finger 2
|
|
chr2_-_169573766
Show fit
|
0.33 |
ENST00000438035.5
ENST00000453929.6
|
FASTKD1
|
FAST kinase domains 1
|
|
chr5_+_40841308
Show fit
|
0.32 |
ENST00000381677.4
ENST00000254691.10
|
CARD6
|
caspase recruitment domain family member 6
|
|
chr11_+_59787067
Show fit
|
0.32 |
ENST00000528805.1
|
STX3
|
syntaxin 3
|
|
chr1_-_111200633
Show fit
|
0.32 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D
|
|
chrX_-_112679919
Show fit
|
0.31 |
ENST00000371968.8
|
LHFPL1
|
LHFPL tetraspan subfamily member 1
|
|
chr3_+_52777580
Show fit
|
0.30 |
ENST00000273283.7
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr6_+_52420992
Show fit
|
0.30 |
ENST00000636954.1
ENST00000636566.1
ENST00000638075.1
|
EFHC1
|
EF-hand domain containing 1
|
|
chr1_-_11047225
Show fit
|
0.30 |
ENST00000400898.3
ENST00000400897.8
|
MASP2
|
mannan binding lectin serine peptidase 2
|
|
chr1_+_220528112
Show fit
|
0.30 |
ENST00000366917.6
ENST00000402574.5
ENST00000611084.4
ENST00000366918.8
|
MARK1
|
microtubule affinity regulating kinase 1
|
|
chr22_+_26621952
Show fit
|
0.29 |
ENST00000354760.4
|
CRYBA4
|
crystallin beta A4
|
|
chr2_+_47403116
Show fit
|
0.29 |
ENST00000645506.1
ENST00000406134.5
ENST00000233146.7
|
MSH2
|
mutS homolog 2
|
|
chr4_+_85475167
Show fit
|
0.29 |
ENST00000503995.5
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr9_-_120477354
Show fit
|
0.29 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2
|
|
chr2_+_47403061
Show fit
|
0.29 |
ENST00000543555.6
|
MSH2
|
mutS homolog 2
|
|
chr10_+_101131284
Show fit
|
0.28 |
ENST00000370196.11
ENST00000467928.2
|
TLX1
|
T cell leukemia homeobox 1
|
|
chr9_-_28670285
Show fit
|
0.28 |
ENST00000379992.6
ENST00000308675.5
ENST00000613945.3
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr11_-_102530738
Show fit
|
0.28 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7
|
|
chrX_+_136532205
Show fit
|
0.28 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1
|
|
chr17_-_19748355
Show fit
|
0.28 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr7_+_117014881
Show fit
|
0.28 |
ENST00000422922.5
ENST00000432298.5
|
ST7
|
suppression of tumorigenicity 7
|
|
chr9_+_79572572
Show fit
|
0.26 |
ENST00000435650.5
ENST00000414465.5
ENST00000376537.8
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr22_+_35066136
Show fit
|
0.26 |
ENST00000308700.6
ENST00000404699.7
|
ISX
|
intestine specific homeobox
|
|
chr17_+_38717424
Show fit
|
0.26 |
ENST00000615858.1
|
MLLT6
|
MLLT6, PHD finger containing
|
|
chr6_+_36676489
Show fit
|
0.26 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A
|
|
chr6_+_156776020
Show fit
|
0.26 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B
|
|
chr9_+_79572715
Show fit
|
0.26 |
ENST00000265284.10
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr1_-_26913964
Show fit
|
0.25 |
ENST00000254227.4
|
NR0B2
|
nuclear receptor subfamily 0 group B member 2
|
|
chr1_-_246566238
Show fit
|
0.25 |
ENST00000366514.5
|
TFB2M
|
transcription factor B2, mitochondrial
|
|
chr8_+_94895837
Show fit
|
0.24 |
ENST00000519136.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6
|
|
chr14_-_31457417
Show fit
|
0.24 |
ENST00000356180.4
|
DTD2
|
D-aminoacyl-tRNA deacylase 2
|
|
chr10_-_79560386
Show fit
|
0.23 |
ENST00000372327.9
ENST00000417041.1
ENST00000640627.1
ENST00000372325.7
|
SFTPA2
|
surfactant protein A2
|
|
chr20_-_44521989
Show fit
|
0.23 |
ENST00000342374.5
ENST00000255175.5
|
SERINC3
|
serine incorporator 3
|
|
chr19_+_18386150
Show fit
|
0.23 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15
|
|
chr6_-_87095059
Show fit
|
0.22 |
ENST00000369582.6
ENST00000610310.3
ENST00000630630.2
ENST00000627148.3
ENST00000625577.1
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr3_+_142596385
Show fit
|
0.22 |
ENST00000457734.7
ENST00000483373.5
ENST00000475296.5
ENST00000495744.5
ENST00000476044.5
ENST00000461644.5
ENST00000464320.5
|
PLS1
|
plastin 1
|
|
chr1_-_244860376
Show fit
|
0.22 |
ENST00000638716.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr12_-_68253502
Show fit
|
0.21 |
ENST00000328087.6
ENST00000538666.6
|
IL22
|
interleukin 22
|
|
chr1_-_216423396
Show fit
|
0.21 |
ENST00000366942.3
ENST00000674083.1
ENST00000307340.8
|
USH2A
|
usherin
|
|
chr7_+_135148041
Show fit
|
0.20 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140
|
|
chr14_-_99272184
Show fit
|
0.20 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B
|
|
chr10_-_50279715
Show fit
|
0.20 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2
|
|
chr19_-_18791297
Show fit
|
0.19 |
ENST00000542601.6
ENST00000222271.7
ENST00000425807.1
|
COMP
|
cartilage oligomeric matrix protein
|
|
chr1_-_203351115
Show fit
|
0.19 |
ENST00000354955.5
|
FMOD
|
fibromodulin
|
|
chr6_-_159045104
Show fit
|
0.19 |
ENST00000326965.7
|
TAGAP
|
T cell activation RhoGTPase activating protein
|
|
chr14_-_99271485
Show fit
|
0.19 |
ENST00000345514.2
ENST00000443726.2
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B
|
|
chr10_+_102776237
Show fit
|
0.19 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like
|
|
chr9_+_137230757
Show fit
|
0.18 |
ENST00000673865.1
ENST00000538474.5
ENST00000673835.1
ENST00000673953.1
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 member 3
|
|
chr20_-_47355657
Show fit
|
0.17 |
ENST00000311275.11
|
ZMYND8
|
zinc finger MYND-type containing 8
|
|
chr6_-_106629472
Show fit
|
0.17 |
ENST00000369063.8
|
RTN4IP1
|
reticulon 4 interacting protein 1
|
|
chr3_+_174859315
Show fit
|
0.17 |
ENST00000454872.6
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase like 2
|
|
chr2_+_237968770
Show fit
|
0.17 |
ENST00000434655.5
ENST00000612130.4
|
UBE2F
|
ubiquitin conjugating enzyme E2 F (putative)
|
|
chr3_-_120349294
Show fit
|
0.17 |
ENST00000295628.4
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr7_+_114416286
Show fit
|
0.17 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2
|
|
chr20_+_33031648
Show fit
|
0.16 |
ENST00000349552.1
|
BPIFB6
|
BPI fold containing family B member 6
|
|
chr19_-_49877279
Show fit
|
0.16 |
ENST00000391832.7
ENST00000344175.10
|
AKT1S1
|
AKT1 substrate 1
|
|
chr9_-_95516959
Show fit
|
0.16 |
ENST00000437951.6
ENST00000430669.6
ENST00000468211.6
|
PTCH1
|
patched 1
|
|
chr19_+_38390055
Show fit
|
0.16 |
ENST00000587947.5
ENST00000338502.8
|
SPRED3
|
sprouty related EVH1 domain containing 3
|
|
chr2_+_151410090
Show fit
|
0.16 |
ENST00000430328.6
|
RIF1
|
replication timing regulatory factor 1
|
|
chr10_+_79610932
Show fit
|
0.15 |
ENST00000428376.6
ENST00000398636.8
ENST00000419470.6
ENST00000429958.5
|
SFTPA1
|
surfactant protein A1
|
|
chr9_+_79571956
Show fit
|
0.15 |
ENST00000376552.8
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr10_+_113709261
Show fit
|
0.15 |
ENST00000672138.1
ENST00000452490.3
|
CASP7
|
caspase 7
|
|
chr6_-_41747390
Show fit
|
0.15 |
ENST00000356667.8
ENST00000373025.7
ENST00000425343.6
|
PGC
|
progastricsin
|
|
chr10_+_89332484
Show fit
|
0.15 |
ENST00000371811.4
ENST00000680037.1
ENST00000679583.1
ENST00000679897.1
|
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3
|
|
chr1_+_174964750
Show fit
|
0.14 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1 like
|
|
chr5_-_142686079
Show fit
|
0.14 |
ENST00000337706.7
|
FGF1
|
fibroblast growth factor 1
|
|
chr22_+_20774092
Show fit
|
0.13 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1
|
|
chr18_-_55403682
Show fit
|
0.13 |
ENST00000564228.5
ENST00000630828.2
|
TCF4
|
transcription factor 4
|
|
chr4_-_69495897
Show fit
|
0.13 |
ENST00000305107.7
ENST00000639621.1
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4
|
|
chr2_-_182427014
Show fit
|
0.13 |
ENST00000409365.5
ENST00000351439.9
|
PDE1A
|
phosphodiesterase 1A
|
|
chr3_+_52211442
Show fit
|
0.13 |
ENST00000459884.1
|
ALAS1
|
5'-aminolevulinate synthase 1
|
|
chr1_-_43389768
Show fit
|
0.12 |
ENST00000372455.4
ENST00000372457.9
ENST00000290663.10
|
MED8
|
mediator complex subunit 8
|
|
chr4_-_69495861
Show fit
|
0.12 |
ENST00000512583.5
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4
|
|
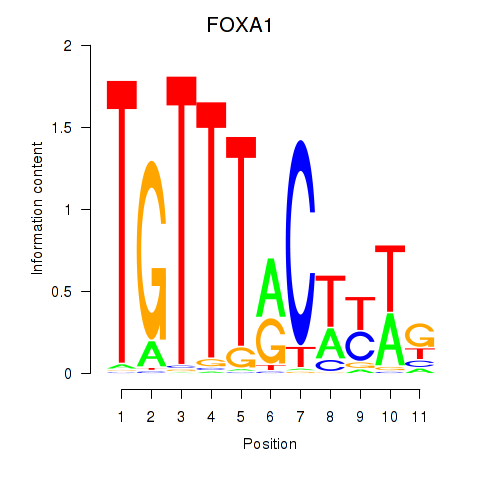
chr14_-_37595224
Show fit
|
0.12 |
ENST00000250448.5
|
FOXA1
|
forkhead box A1
|
|
chr17_-_42745025
Show fit
|
0.12 |
ENST00000592492.5
ENST00000585893.5
ENST00000593214.5
ENST00000590078.5
ENST00000428826.7
ENST00000586382.5
ENST00000415827.6
ENST00000592743.5
ENST00000586089.5
|
EZH1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit
|
|
chr3_-_190449782
Show fit
|
0.12 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207
|
|
chrX_-_124963768
Show fit
|
0.12 |
ENST00000371130.7
ENST00000422452.2
|
TENM1
|
teneurin transmembrane protein 1
|
|
chr14_+_55661272
Show fit
|
0.12 |
ENST00000555573.5
|
KTN1
|
kinectin 1
|
|
chr18_-_54959391
Show fit
|
0.11 |
ENST00000591504.6
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr1_+_43389889
Show fit
|
0.11 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr17_-_17582417
Show fit
|
0.11 |
ENST00000395783.5
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr14_-_36582593
Show fit
|
0.11 |
ENST00000258829.6
|
NKX2-8
|
NK2 homeobox 8
|
|
chr11_+_31812307
Show fit
|
0.11 |
ENST00000643436.1
ENST00000646959.1
ENST00000645942.1
ENST00000530348.5
|
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA
novel protein
|
|
chr6_-_56843153
Show fit
|
0.11 |
ENST00000361203.7
ENST00000523817.1
|
DST
|
dystonin
|
|
chr13_+_75760362
Show fit
|
0.11 |
ENST00000534657.5
|
LMO7
|
LIM domain 7
|
|
chr14_+_64504743
Show fit
|
0.10 |
ENST00000683701.1
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr13_-_99258366
Show fit
|
0.10 |
ENST00000397470.5
ENST00000397473.7
|
GPR18
|
G protein-coupled receptor 18
|
|
chr7_-_81770122
Show fit
|
0.10 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor
|
|
chr5_-_41510623
Show fit
|
0.10 |
ENST00000328457.5
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3
|
|
chr6_+_143608170
Show fit
|
0.09 |
ENST00000427704.6
ENST00000305766.10
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chrX_+_1268807
Show fit
|
0.09 |
ENST00000381524.8
ENST00000381529.9
ENST00000412290.6
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr14_-_31457495
Show fit
|
0.09 |
ENST00000310850.9
|
DTD2
|
D-aminoacyl-tRNA deacylase 2
|
|
chr3_+_122183664
Show fit
|
0.09 |
ENST00000639785.2
|
CASR
|
calcium sensing receptor
|
|
chr20_+_64164446
Show fit
|
0.09 |
ENST00000328439.6
|
MYT1
|
myelin transcription factor 1
|
|
chr2_+_102473219
Show fit
|
0.09 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4
|
|
chr5_-_41510554
Show fit
|
0.09 |
ENST00000377801.8
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3
|
|
chr11_-_124310837
Show fit
|
0.09 |
ENST00000357821.2
|
OR8D1
|
olfactory receptor family 8 subfamily D member 1
|
|
chr8_+_125430333
Show fit
|
0.08 |
ENST00000311922.4
|
TRIB1
|
tribbles pseudokinase 1
|
|
chr11_-_107712049
Show fit
|
0.08 |
ENST00000305991.3
|
SLN
|
sarcolipin
|
|
chr2_+_151409878
Show fit
|
0.08 |
ENST00000453091.6
ENST00000428287.6
ENST00000444746.7
ENST00000243326.9
ENST00000414861.6
|
RIF1
|
replication timing regulatory factor 1
|
|
chr20_+_64164474
Show fit
|
0.08 |
ENST00000622439.4
ENST00000536311.5
|
MYT1
|
myelin transcription factor 1
|
|
chr12_-_120327762
Show fit
|
0.08 |
ENST00000308366.9
ENST00000423423.3
|
PLA2G1B
|
phospholipase A2 group IB
|
|
chr20_+_64164566
Show fit
|
0.08 |
ENST00000650655.1
|
MYT1
|
myelin transcription factor 1
|
|
chr1_-_178871022
Show fit
|
0.08 |
ENST00000367629.1
|
ANGPTL1
|
angiopoietin like 1
|
|
chrX_-_15384402
Show fit
|
0.07 |
ENST00000297904.4
|
VEGFD
|
vascular endothelial growth factor D
|
|
chr17_-_41315706
Show fit
|
0.07 |
ENST00000334202.5
|
KRTAP17-1
|
keratin associated protein 17-1
|
|
chr1_-_178871060
Show fit
|
0.07 |
ENST00000234816.7
|
ANGPTL1
|
angiopoietin like 1
|
|
chr10_-_125160499
Show fit
|
0.07 |
ENST00000494626.6
ENST00000337195.9
|
CTBP2
|
C-terminal binding protein 2
|
|
chr12_-_21334858
Show fit
|
0.07 |
ENST00000445053.1
ENST00000458504.5
ENST00000422327.5
ENST00000683939.1
|
SLCO1A2
|
solute carrier organic anion transporter family member 1A2
|
|
chr16_+_4846652
Show fit
|
0.07 |
ENST00000592120.5
|
UBN1
|
ubinuclein 1
|
|
chr11_+_121102666
Show fit
|
0.07 |
ENST00000264037.2
|
TECTA
|
tectorin alpha
|
|
chr3_+_138010143
Show fit
|
0.07 |
ENST00000183605.10
|
CLDN18
|
claudin 18
|
|
chr16_-_48610150
Show fit
|
0.07 |
ENST00000262384.4
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr5_+_162068031
Show fit
|
0.07 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr12_-_48004496
Show fit
|
0.06 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain
|
|
chr11_+_1157946
Show fit
|
0.06 |
ENST00000621226.2
|
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming
|
|
chr10_-_48274567
Show fit
|
0.06 |
ENST00000636244.1
ENST00000374201.8
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr12_+_10505890
Show fit
|
0.06 |
ENST00000538173.1
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B
|
|
chr5_+_162067500
Show fit
|
0.06 |
ENST00000639384.1
ENST00000640985.1
ENST00000638772.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr7_+_73828160
Show fit
|
0.06 |
ENST00000431918.1
|
CLDN4
|
claudin 4
|