Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for FOXA3_FOXC2
Z-value: 0.35
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
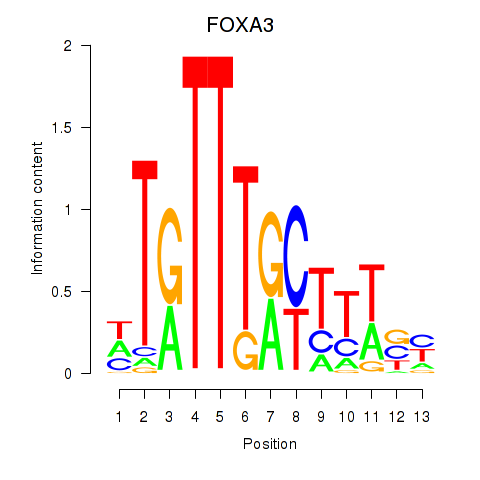
FOXA3
|
ENSG00000170608.3 | FOXA3 |
|
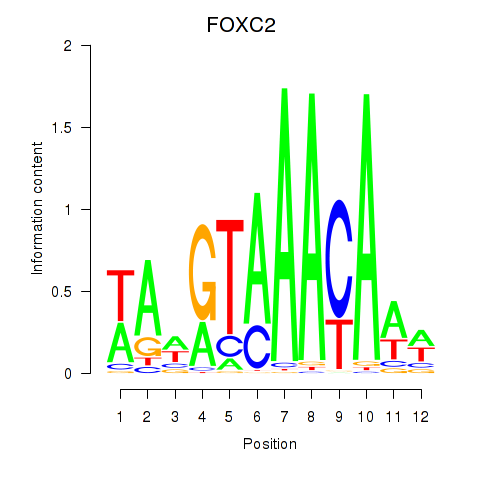
FOXC2
|
ENSG00000176692.8 | FOXC2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC2 | hg38_v1_chr16_+_86566821_86566836 | 0.09 | 6.3e-01 | Click! |
| FOXA3 | hg38_v1_chr19_+_45864318_45864334 | 0.08 | 6.6e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33235987 | 2.91 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr13_+_50015438 | 1.21 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr13_+_50015254 | 1.19 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr6_+_52420992 | 1.15 |
ENST00000636954.1
ENST00000636566.1 ENST00000638075.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr4_-_88284747 | 1.08 |
ENST00000514204.1
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr11_+_27055215 | 0.98 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr20_+_33283205 | 0.93 |
ENST00000253354.2
|
BPIFB1
|
BPI fold containing family B member 1 |
| chr11_-_108593738 | 0.80 |
ENST00000525344.5
ENST00000265843.9 |
EXPH5
|
exophilin 5 |
| chr13_-_38990824 | 0.77 |
ENST00000379631.9
|
STOML3
|
stomatin like 3 |
| chr13_-_38990856 | 0.76 |
ENST00000423210.1
|
STOML3
|
stomatin like 3 |
| chr7_+_143132069 | 0.70 |
ENST00000291009.4
|
PIP
|
prolactin induced protein |
| chr1_-_206921987 | 0.61 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr13_-_35855627 | 0.57 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr4_+_164754045 | 0.55 |
ENST00000515485.5
|
SMIM31
|
small integral membrane protein 31 |
| chr20_+_9069076 | 0.52 |
ENST00000378473.9
|
PLCB4
|
phospholipase C beta 4 |
| chr1_+_226940279 | 0.51 |
ENST00000366778.5
ENST00000366777.4 |
COQ8A
|
coenzyme Q8A |
| chr1_+_74235377 | 0.49 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_-_206921867 | 0.46 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr3_-_108222362 | 0.45 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 |
| chr9_-_4300049 | 0.45 |
ENST00000381971.8
|
GLIS3
|
GLIS family zinc finger 3 |
| chr11_+_6876625 | 0.42 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor family 10 subfamily A member 4 |
| chr4_+_164754116 | 0.41 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr12_-_71157872 | 0.41 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr14_+_104801082 | 0.40 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr13_-_85799400 | 0.40 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr3_-_45915698 | 0.38 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr9_-_72953047 | 0.35 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chr15_+_70892809 | 0.35 |
ENST00000260382.10
ENST00000560755.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_+_12746192 | 0.32 |
ENST00000614859.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr12_-_76486061 | 0.32 |
ENST00000548341.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr10_+_94089034 | 0.32 |
ENST00000676102.1
ENST00000371385.8 |
PLCE1
|
phospholipase C epsilon 1 |
| chr17_+_9021501 | 0.32 |
ENST00000173229.7
|
NTN1
|
netrin 1 |
| chr10_+_94089067 | 0.32 |
ENST00000371375.1
ENST00000675218.1 |
PLCE1
|
phospholipase C epsilon 1 |
| chr3_-_108222383 | 0.30 |
ENST00000264538.4
|
IFT57
|
intraflagellar transport 57 |
| chr18_+_3449413 | 0.30 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr7_-_138627444 | 0.29 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr17_-_31314066 | 0.28 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr1_-_201127184 | 0.27 |
ENST00000449188.3
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr19_-_45584810 | 0.27 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr2_-_174846405 | 0.26 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr13_-_35855758 | 0.25 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr3_+_69936583 | 0.25 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr17_+_4710622 | 0.25 |
ENST00000574954.5
ENST00000269260.7 ENST00000346341.6 ENST00000572457.5 ENST00000381488.10 ENST00000412477.7 ENST00000571428.5 ENST00000575877.5 |
ARRB2
|
arrestin beta 2 |
| chr12_-_49187369 | 0.25 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr9_-_20382461 | 0.24 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr1_-_56966133 | 0.24 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr7_+_117014881 | 0.24 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr9_-_92424427 | 0.23 |
ENST00000375550.5
|
OMD
|
osteomodulin |
| chr14_+_56118404 | 0.22 |
ENST00000267460.9
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr1_+_87331668 | 0.21 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr12_-_39340963 | 0.21 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A |
| chr22_-_30471986 | 0.21 |
ENST00000401751.5
ENST00000402286.5 ENST00000403066.5 ENST00000215812.9 |
SEC14L3
|
SEC14 like lipid binding 3 |
| chr1_+_18631006 | 0.21 |
ENST00000375375.7
|
PAX7
|
paired box 7 |
| chr1_-_56966006 | 0.21 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr17_-_7043906 | 0.20 |
ENST00000308009.5
ENST00000447225.1 |
SLC16A11
|
solute carrier family 16 member 11 |
| chr17_-_7044091 | 0.20 |
ENST00000574600.3
ENST00000662352.3 ENST00000673828.2 |
SLC16A11
|
solute carrier family 16 member 11 |
| chr18_+_63907948 | 0.20 |
ENST00000238508.8
|
SERPINB10
|
serpin family B member 10 |
| chr1_+_18631513 | 0.19 |
ENST00000400661.3
|
PAX7
|
paired box 7 |
| chr5_+_90640718 | 0.19 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr3_+_171843337 | 0.19 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr1_+_244051275 | 0.19 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr10_-_48274567 | 0.19 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr3_-_149792547 | 0.19 |
ENST00000446160.7
ENST00000462519.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr17_-_28368012 | 0.19 |
ENST00000555059.2
|
ENSG00000273171.1
|
novel protein, readthrough between VTN and SEBOX |
| chr2_+_127418420 | 0.18 |
ENST00000234071.8
ENST00000429925.5 ENST00000442644.5 |
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr17_+_7583828 | 0.18 |
ENST00000396501.8
ENST00000250124.11 ENST00000584378.5 ENST00000423172.6 ENST00000579445.5 ENST00000585217.5 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr1_+_61082553 | 0.18 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr7_+_114414809 | 0.18 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr17_-_28406160 | 0.17 |
ENST00000618626.1
ENST00000612814.5 |
SLC46A1
|
solute carrier family 46 member 1 |
| chr7_+_135148041 | 0.17 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chrX_+_136648214 | 0.17 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr18_-_28036585 | 0.16 |
ENST00000399380.7
|
CDH2
|
cadherin 2 |
| chr19_+_42302098 | 0.16 |
ENST00000598490.1
ENST00000341747.8 |
PRR19
|
proline rich 19 |
| chr22_+_35066136 | 0.16 |
ENST00000308700.6
ENST00000404699.7 |
ISX
|
intestine specific homeobox |
| chr16_+_14708944 | 0.16 |
ENST00000526520.5
ENST00000531598.6 |
NPIPA3
|
nuclear pore complex interacting protein family member A3 |
| chr7_-_141973773 | 0.16 |
ENST00000547270.1
|
TAS2R38
|
taste 2 receptor member 38 |
| chrX_+_136648138 | 0.16 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr2_-_174847015 | 0.15 |
ENST00000650938.1
|
CHN1
|
chimerin 1 |
| chr1_+_113929304 | 0.15 |
ENST00000426820.7
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr8_-_132760548 | 0.15 |
ENST00000519187.5
ENST00000523829.5 ENST00000677595.1 ENST00000356838.7 ENST00000377901.8 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr14_-_25010604 | 0.15 |
ENST00000550887.5
|
STXBP6
|
syntaxin binding protein 6 |
| chr14_+_20688756 | 0.15 |
ENST00000397990.5
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin ribonuclease A family member 4 |
| chr4_+_85475131 | 0.14 |
ENST00000395184.6
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_-_26056460 | 0.14 |
ENST00000343677.4
|
H1-2
|
H1.2 linker histone, cluster member |
| chr5_+_162068031 | 0.14 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr6_+_44342684 | 0.13 |
ENST00000288390.2
|
SPATS1
|
spermatogenesis associated serine rich 1 |
| chr11_-_85686123 | 0.13 |
ENST00000316398.5
|
CCDC89
|
coiled-coil domain containing 89 |
| chr5_+_162067500 | 0.13 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr19_-_18791297 | 0.13 |
ENST00000542601.6
ENST00000222271.7 ENST00000425807.1 |
COMP
|
cartilage oligomeric matrix protein |
| chr5_-_41213505 | 0.13 |
ENST00000337836.10
ENST00000433294.1 |
C6
|
complement C6 |
| chr5_-_116536458 | 0.13 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A |
| chr18_+_75210789 | 0.13 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr6_+_44342639 | 0.12 |
ENST00000674044.1
ENST00000515220.5 ENST00000323108.12 |
SPATS1
|
spermatogenesis associated serine rich 1 |
| chr5_+_162067858 | 0.12 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr1_+_18630839 | 0.12 |
ENST00000420770.7
|
PAX7
|
paired box 7 |
| chr10_-_124093582 | 0.12 |
ENST00000462406.1
ENST00000435907.6 |
CHST15
|
carbohydrate sulfotransferase 15 |
| chr1_+_40396766 | 0.12 |
ENST00000539317.2
|
SMAP2
|
small ArfGAP2 |
| chr17_+_74431338 | 0.12 |
ENST00000342648.9
ENST00000652232.1 ENST00000481232.2 |
GPRC5C
|
G protein-coupled receptor class C group 5 member C |
| chr6_+_156776020 | 0.11 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B |
| chr1_-_150697128 | 0.11 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr6_-_132659178 | 0.11 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr15_+_96325935 | 0.11 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr11_-_116837586 | 0.11 |
ENST00000375320.5
ENST00000359492.6 ENST00000375329.6 ENST00000375323.5 ENST00000236850.5 |
APOA1
|
apolipoprotein A1 |
| chr3_+_132597260 | 0.11 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr5_+_162067764 | 0.11 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_40841308 | 0.11 |
ENST00000381677.4
ENST00000254691.10 |
CARD6
|
caspase recruitment domain family member 6 |
| chr10_-_50279715 | 0.11 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chr9_+_68241854 | 0.10 |
ENST00000616550.4
ENST00000618217.4 ENST00000377342.9 ENST00000478048.5 ENST00000360171.11 |
CBWD3
|
COBW domain containing 3 |
| chr9_-_41189310 | 0.10 |
ENST00000456520.5
ENST00000377391.8 ENST00000613716.4 ENST00000617933.1 |
CBWD6
|
COBW domain containing 6 |
| chr12_-_56934403 | 0.10 |
ENST00000293502.2
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C member 7 |
| chr16_+_69924984 | 0.10 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_+_85475167 | 0.10 |
ENST00000503995.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr7_+_151341764 | 0.10 |
ENST00000413040.7
ENST00000470229.6 ENST00000568733.6 |
NUB1
|
negative regulator of ubiquitin like proteins 1 |
| chr18_-_55403682 | 0.10 |
ENST00000564228.5
ENST00000630828.2 |
TCF4
|
transcription factor 4 |
| chr16_+_20764036 | 0.10 |
ENST00000440284.6
|
ACSM3
|
acyl-CoA synthetase medium chain family member 3 |
| chr5_+_141408000 | 0.10 |
ENST00000616430.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr14_-_31457495 | 0.10 |
ENST00000310850.9
|
DTD2
|
D-aminoacyl-tRNA deacylase 2 |
| chr5_+_140827950 | 0.09 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr8_+_19313685 | 0.09 |
ENST00000265807.8
ENST00000518040.5 |
SH2D4A
|
SH2 domain containing 4A |
| chr3_-_114624193 | 0.09 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr14_+_74084947 | 0.09 |
ENST00000674221.1
ENST00000554938.2 |
LIN52
|
lin-52 DREAM MuvB core complex component |
| chr1_-_246566238 | 0.09 |
ENST00000366514.5
|
TFB2M
|
transcription factor B2, mitochondrial |
| chr17_-_31314040 | 0.08 |
ENST00000330927.5
|
EVI2B
|
ecotropic viral integration site 2B |
| chr4_+_164877164 | 0.08 |
ENST00000507152.6
ENST00000515275.1 |
APELA
|
apelin receptor early endogenous ligand |
| chr5_-_59276109 | 0.08 |
ENST00000503258.5
|
PDE4D
|
phosphodiesterase 4D |
| chr1_-_167553799 | 0.08 |
ENST00000466652.2
|
CREG1
|
cellular repressor of E1A stimulated genes 1 |
| chr2_+_113437691 | 0.08 |
ENST00000259199.9
ENST00000416503.6 ENST00000433343.6 |
CBWD2
|
COBW domain containing 2 |
| chr1_-_169586471 | 0.08 |
ENST00000367797.9
|
F5
|
coagulation factor V |
| chr2_+_227616998 | 0.08 |
ENST00000641801.1
|
SCYGR4
|
small cysteine and glycine repeat containing 4 |
| chr19_-_46654657 | 0.08 |
ENST00000300875.4
|
DACT3
|
dishevelled binding antagonist of beta catenin 3 |
| chr14_-_31457417 | 0.08 |
ENST00000356180.4
|
DTD2
|
D-aminoacyl-tRNA deacylase 2 |
| chr10_-_20897288 | 0.08 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr13_+_52024691 | 0.08 |
ENST00000521776.2
|
UTP14C
|
UTP14C small subunit processome component |
| chr2_+_87748087 | 0.08 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr22_+_25111810 | 0.07 |
ENST00000637069.1
|
KIAA1671
|
KIAA1671 |
| chr14_+_55661272 | 0.07 |
ENST00000555573.5
|
KTN1
|
kinectin 1 |
| chrX_-_81201886 | 0.07 |
ENST00000451455.1
ENST00000358130.7 ENST00000436386.5 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr2_+_46617180 | 0.07 |
ENST00000238892.4
|
CRIPT
|
CXXC repeat containing interactor of PDZ3 domain |
| chr1_-_244860376 | 0.07 |
ENST00000638716.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U |
| chr5_+_62578810 | 0.07 |
ENST00000334994.6
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr1_+_174964750 | 0.07 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr3_-_187737943 | 0.07 |
ENST00000438077.1
|
BCL6
|
BCL6 transcription repressor |
| chr12_-_47705971 | 0.07 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr20_+_11917859 | 0.07 |
ENST00000618296.4
ENST00000378226.7 |
BTBD3
|
BTB domain containing 3 |
| chr4_+_183905266 | 0.07 |
ENST00000308497.9
|
STOX2
|
storkhead box 2 |
| chr1_+_205256189 | 0.07 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr4_-_73620629 | 0.07 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr1_-_151459471 | 0.07 |
ENST00000271715.7
|
POGZ
|
pogo transposable element derived with ZNF domain |
| chr3_+_35679690 | 0.07 |
ENST00000413378.5
ENST00000417925.5 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr15_+_60004305 | 0.07 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1 |
| chr19_-_45584769 | 0.07 |
ENST00000263275.5
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr1_+_113929350 | 0.06 |
ENST00000369559.8
ENST00000626993.2 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr3_+_137998735 | 0.06 |
ENST00000343735.8
|
CLDN18
|
claudin 18 |
| chr5_+_162067458 | 0.06 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr1_-_53940100 | 0.06 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr6_+_20534441 | 0.06 |
ENST00000274695.8
ENST00000613575.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1 like 1 |
| chr12_-_47705990 | 0.06 |
ENST00000432584.7
ENST00000005386.8 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr10_-_125160499 | 0.06 |
ENST00000494626.6
ENST00000337195.9 |
CTBP2
|
C-terminal binding protein 2 |
| chr1_-_151459169 | 0.06 |
ENST00000368863.6
ENST00000409503.5 ENST00000491586.5 ENST00000533351.5 |
POGZ
|
pogo transposable element derived with ZNF domain |
| chr3_+_138010143 | 0.06 |
ENST00000183605.10
|
CLDN18
|
claudin 18 |
| chr8_+_74824526 | 0.06 |
ENST00000649643.1
ENST00000260113.7 |
PI15
|
peptidase inhibitor 15 |
| chr4_-_139302460 | 0.06 |
ENST00000394223.2
ENST00000676245.1 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr4_+_74308463 | 0.06 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr8_-_119638780 | 0.06 |
ENST00000522826.5
ENST00000520066.5 ENST00000259486.10 ENST00000075322.11 |
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr6_-_87095059 | 0.06 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chrX_+_10156960 | 0.06 |
ENST00000380833.9
|
CLCN4
|
chloride voltage-gated channel 4 |
| chr4_+_145482761 | 0.06 |
ENST00000507367.1
ENST00000394092.6 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr3_+_155870623 | 0.06 |
ENST00000295920.7
ENST00000496455.7 |
GMPS
|
guanine monophosphate synthase |
| chr1_+_246566422 | 0.06 |
ENST00000366513.9
ENST00000366512.7 |
CNST
|
consortin, connexin sorting protein |
| chr2_+_134838610 | 0.05 |
ENST00000356140.10
ENST00000392928.5 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr4_-_139302516 | 0.05 |
ENST00000394228.5
ENST00000539387.5 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr6_+_52186373 | 0.05 |
ENST00000648244.1
|
IL17A
|
interleukin 17A |
| chr10_-_125161019 | 0.05 |
ENST00000411419.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr12_-_121580441 | 0.05 |
ENST00000377069.8
|
KDM2B
|
lysine demethylase 2B |
| chr10_+_122163672 | 0.05 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr21_-_42366525 | 0.05 |
ENST00000291527.3
|
TFF1
|
trefoil factor 1 |
| chr9_+_137230757 | 0.05 |
ENST00000673865.1
ENST00000538474.5 ENST00000673835.1 ENST00000673953.1 ENST00000361134.2 |
SLC34A3
|
solute carrier family 34 member 3 |
| chr1_+_43389874 | 0.05 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr2_+_68365274 | 0.05 |
ENST00000234313.8
|
PLEK
|
pleckstrin |
| chr14_-_100569780 | 0.05 |
ENST00000355173.7
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr10_-_30999469 | 0.05 |
ENST00000538351.6
|
ZNF438
|
zinc finger protein 438 |
| chr5_+_98769273 | 0.05 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr14_-_24195334 | 0.05 |
ENST00000530563.1
ENST00000528895.5 ENST00000528669.5 ENST00000532632.1 ENST00000261789.9 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chr10_-_69416912 | 0.05 |
ENST00000373306.5
|
TACR2
|
tachykinin receptor 2 |
| chr6_-_168079159 | 0.05 |
ENST00000283309.10
|
FRMD1
|
FERM domain containing 1 |
| chr17_+_42998379 | 0.05 |
ENST00000253788.12
ENST00000589913.6 |
RPL27
|
ribosomal protein L27 |
| chr5_+_62412755 | 0.05 |
ENST00000325324.11
|
IPO11
|
importin 11 |
| chr5_+_141408032 | 0.04 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr7_-_81770122 | 0.04 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr6_+_10528326 | 0.04 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr2_+_168802610 | 0.04 |
ENST00000397206.6
ENST00000317647.12 ENST00000397209.6 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr9_+_65675834 | 0.04 |
ENST00000377392.9
ENST00000377384.5 ENST00000430059.6 ENST00000429800.6 ENST00000382405.8 ENST00000377395.8 |
CBWD5
|
COBW domain containing 5 |
| chr1_-_169586539 | 0.04 |
ENST00000367796.3
|
F5
|
coagulation factor V |
| chr12_+_57694118 | 0.04 |
ENST00000315970.12
ENST00000547079.5 ENST00000439210.6 ENST00000389146.10 ENST00000413095.6 ENST00000551035.5 ENST00000257966.12 ENST00000435406.6 ENST00000550372.5 ENST00000389142.9 |
OS9
|
OS9 endoplasmic reticulum lectin |
| chr1_+_222928415 | 0.04 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr6_+_26251607 | 0.04 |
ENST00000619466.2
|
H2BC9
|
H2B clustered histone 9 |
| chr15_+_75043263 | 0.04 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr6_-_41747390 | 0.04 |
ENST00000356667.8
ENST00000373025.7 ENST00000425343.6 |
PGC
|
progastricsin |
| chr14_-_21094488 | 0.04 |
ENST00000555270.5
|
ZNF219
|
zinc finger protein 219 |
| chr4_-_99144238 | 0.04 |
ENST00000512499.5
ENST00000504125.1 ENST00000505590.5 ENST00000629236.2 ENST00000508393.5 ENST00000265512.12 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.2 | 0.5 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.1 | 2.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.4 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.9 | GO:0034144 | innate immune response in mucosa(GO:0002227) negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.4 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 1.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.2 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.7 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.8 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.1 | GO:0021678 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.0 | 0.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.9 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 1.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.0 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |