Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
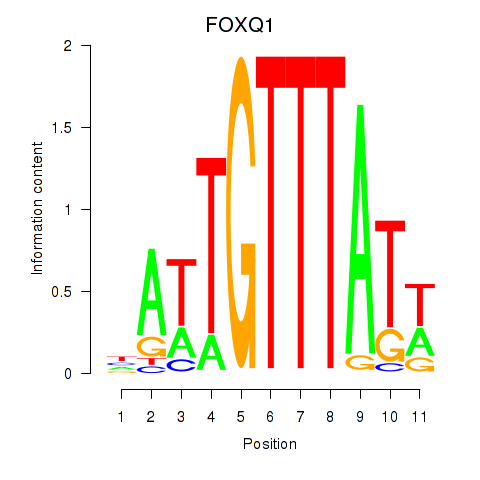
Results for FOXQ1
Z-value: 0.83
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.7 | FOXQ1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg38_v1_chr6_+_1312090_1312105 | 0.62 | 2.4e-04 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_131637296 | 5.08 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr12_-_71157872 | 4.92 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr9_+_69820799 | 4.24 |
ENST00000377197.8
|
C9orf135
|
chromosome 9 open reading frame 135 |
| chr9_+_122159886 | 3.74 |
ENST00000373764.8
ENST00000536616.5 |
MORN5
|
MORN repeat containing 5 |
| chr13_-_38990824 | 3.74 |
ENST00000379631.9
|
STOML3
|
stomatin like 3 |
| chr13_-_38990856 | 3.69 |
ENST00000423210.1
|
STOML3
|
stomatin like 3 |
| chr4_+_164754045 | 3.62 |
ENST00000515485.5
|
SMIM31
|
small integral membrane protein 31 |
| chr3_-_19934189 | 3.58 |
ENST00000295824.14
|
EFHB
|
EF-hand domain family member B |
| chr3_-_167653952 | 3.26 |
ENST00000466760.5
ENST00000479765.5 |
WDR49
|
WD repeat domain 49 |
| chr6_-_52909666 | 3.23 |
ENST00000370968.5
ENST00000211122.4 |
GSTA3
|
glutathione S-transferase alpha 3 |
| chr11_+_72080313 | 3.13 |
ENST00000307198.11
ENST00000538413.6 ENST00000642648.1 ENST00000289488.7 |
ENSG00000284922.2
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chr3_-_167653916 | 3.10 |
ENST00000488012.5
ENST00000682715.1 ENST00000647816.1 |
WDR49
|
WD repeat domain 49 |
| chr11_+_86395166 | 3.09 |
ENST00000528728.1
|
CCDC81
|
coiled-coil domain containing 81 |
| chr14_+_96482982 | 3.06 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr17_+_9576627 | 3.06 |
ENST00000396219.7
ENST00000352665.10 |
CFAP52
|
cilia and flagella associated protein 52 |
| chr1_+_103655760 | 2.68 |
ENST00000370083.9
|
AMY1A
|
amylase alpha 1A |
| chr4_+_164754116 | 2.62 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chrX_-_100410264 | 2.53 |
ENST00000373034.8
|
PCDH19
|
protocadherin 19 |
| chr9_+_69820827 | 2.50 |
ENST00000527647.5
ENST00000480564.1 |
C9orf135
|
chromosome 9 open reading frame 135 |
| chr12_-_71157992 | 2.42 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8 |
| chr15_-_56465130 | 2.41 |
ENST00000260453.4
|
MNS1
|
meiosis specific nuclear structural 1 |
| chr11_+_73950985 | 2.26 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr1_+_103749898 | 2.22 |
ENST00000622339.5
|
AMY1C
|
amylase alpha 1C |
| chr1_+_103617427 | 2.08 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr10_+_125973373 | 1.98 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr1_-_216805367 | 1.91 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr5_+_122129533 | 1.91 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr9_-_35563867 | 1.89 |
ENST00000399742.7
ENST00000619051.4 |
FAM166B
|
family with sequence similarity 166 member B |
| chr11_+_6876625 | 1.83 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor family 10 subfamily A member 4 |
| chr11_-_26572130 | 1.63 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26572102 | 1.59 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr6_-_52803807 | 1.58 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr16_+_4734519 | 1.56 |
ENST00000299320.10
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr16_+_4734457 | 1.53 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr1_+_61077219 | 1.51 |
ENST00000407417.7
|
NFIA
|
nuclear factor I A |
| chr6_-_32666648 | 1.45 |
ENST00000399082.7
ENST00000399079.7 ENST00000374943.8 ENST00000434651.6 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr7_+_92057602 | 1.45 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr7_-_138627444 | 1.45 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr11_-_26572254 | 1.42 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr10_-_73358853 | 1.39 |
ENST00000355577.8
ENST00000310715.7 |
CFAP70
|
cilia and flagella associated protein 70 |
| chr1_+_63523490 | 1.36 |
ENST00000371088.5
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr5_+_141192330 | 1.23 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr3_-_112845950 | 1.22 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr7_+_48171451 | 1.20 |
ENST00000435803.6
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr3_+_171843337 | 1.15 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr3_-_57544324 | 1.14 |
ENST00000495027.5
ENST00000351747.6 ENST00000389536.8 ENST00000311202.7 |
DNAH12
|
dynein axonemal heavy chain 12 |
| chr8_+_93754879 | 1.02 |
ENST00000453906.6
ENST00000683362.1 ENST00000682036.1 ENST00000453321.8 ENST00000409623.8 ENST00000520680.2 ENST00000521517.6 ENST00000452276.6 |
TMEM67
|
transmembrane protein 67 |
| chr6_-_32763522 | 1.02 |
ENST00000435145.6
ENST00000437316.7 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr6_+_38715288 | 1.01 |
ENST00000327475.11
|
DNAH8
|
dynein axonemal heavy chain 8 |
| chr12_-_9999176 | 0.97 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr12_-_49189053 | 0.97 |
ENST00000550767.6
ENST00000546918.1 ENST00000679733.1 ENST00000552924.2 ENST00000301071.12 |
TUBA1A
|
tubulin alpha 1a |
| chr1_-_53940100 | 0.96 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr3_-_114624193 | 0.95 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_60865259 | 0.95 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr6_+_156776020 | 0.90 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B |
| chr6_-_49744434 | 0.89 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr6_-_49744378 | 0.89 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr4_+_39182497 | 0.89 |
ENST00000509560.5
ENST00000512112.5 ENST00000399820.8 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr15_+_50182215 | 0.88 |
ENST00000380902.8
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr16_+_30201057 | 0.88 |
ENST00000569485.5
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr15_+_50182188 | 0.88 |
ENST00000267842.10
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr3_-_114759115 | 0.87 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_-_30571635 | 0.86 |
ENST00000563707.1
ENST00000567855.1 ENST00000223459.11 |
ZNF688
|
zinc finger protein 688 |
| chr1_+_174875505 | 0.84 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr7_-_122995700 | 0.83 |
ENST00000249284.3
|
TAS2R16
|
taste 2 receptor member 16 |
| chr3_+_108602776 | 0.81 |
ENST00000497905.5
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr5_+_73813518 | 0.80 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr6_-_87095059 | 0.79 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr1_+_113929600 | 0.77 |
ENST00000369558.5
ENST00000369561.8 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr14_-_22819721 | 0.75 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr4_+_95051671 | 0.72 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr1_+_171090892 | 0.72 |
ENST00000367755.9
ENST00000479749.1 |
FMO3
|
flavin containing dimethylaniline monoxygenase 3 |
| chr14_+_21030201 | 0.72 |
ENST00000321760.11
ENST00000460647.6 ENST00000530140.6 ENST00000472458.5 |
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr1_-_59926724 | 0.69 |
ENST00000371204.4
|
CYP2J2
|
cytochrome P450 family 2 subfamily J member 2 |
| chr11_+_10455292 | 0.69 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr1_-_149861210 | 0.69 |
ENST00000579512.2
|
H4C15
|
H4 clustered histone 15 |
| chr17_+_45160694 | 0.68 |
ENST00000307275.7
ENST00000585340.2 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chr14_-_89412025 | 0.68 |
ENST00000553840.5
ENST00000556916.5 |
FOXN3
|
forkhead box N3 |
| chr9_-_123184233 | 0.68 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr4_-_185775271 | 0.68 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_123268538 | 0.67 |
ENST00000360998.3
ENST00000348403.10 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chrX_+_106611930 | 0.67 |
ENST00000372544.6
ENST00000372548.9 |
RADX
|
RPA1 related single stranded DNA binding protein, X-linked |
| chr12_-_15221394 | 0.66 |
ENST00000537647.5
ENST00000256953.6 ENST00000546331.5 |
RERG
|
RAS like estrogen regulated growth inhibitor |
| chr2_-_148020689 | 0.66 |
ENST00000457954.5
ENST00000392857.10 ENST00000540442.5 ENST00000535373.5 |
ORC4
|
origin recognition complex subunit 4 |
| chr11_+_112175526 | 0.66 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr16_+_14708944 | 0.64 |
ENST00000526520.5
ENST00000531598.6 |
NPIPA3
|
nuclear pore complex interacting protein family member A3 |
| chr9_-_20382461 | 0.62 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr11_+_121576760 | 0.61 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr18_-_55589770 | 0.61 |
ENST00000565018.6
ENST00000636400.2 |
TCF4
|
transcription factor 4 |
| chr12_+_1629197 | 0.59 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B |
| chr14_+_104801082 | 0.59 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chrX_+_10158448 | 0.59 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr6_-_29045175 | 0.59 |
ENST00000377175.2
|
OR2W1
|
olfactory receptor family 2 subfamily W member 1 |
| chr4_-_139302460 | 0.58 |
ENST00000394223.2
ENST00000676245.1 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr21_+_41370452 | 0.58 |
ENST00000680862.1
|
MX2
|
MX dynamin like GTPase 2 |
| chr16_+_90022600 | 0.58 |
ENST00000620723.4
ENST00000268699.9 |
GAS8
|
growth arrest specific 8 |
| chr11_+_73308237 | 0.57 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr21_-_30216047 | 0.56 |
ENST00000399899.2
|
CLDN8
|
claudin 8 |
| chr1_-_56579555 | 0.55 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr2_+_213284379 | 0.55 |
ENST00000272898.11
ENST00000432529.6 ENST00000413312.5 ENST00000331683.10 ENST00000447990.1 |
SPAG16
|
sperm associated antigen 16 |
| chr14_+_50560137 | 0.53 |
ENST00000358385.12
|
ATL1
|
atlastin GTPase 1 |
| chr3_+_174859315 | 0.52 |
ENST00000454872.6
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase like 2 |
| chr1_-_150697128 | 0.52 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr10_+_70404129 | 0.52 |
ENST00000373218.5
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr1_+_40396766 | 0.52 |
ENST00000539317.2
|
SMAP2
|
small ArfGAP2 |
| chr14_-_60724300 | 0.51 |
ENST00000556952.3
ENST00000216513.5 |
SIX4
|
SIX homeobox 4 |
| chr7_+_2558960 | 0.50 |
ENST00000623361.3
ENST00000402050.7 |
IQCE
|
IQ motif containing E |
| chr17_+_75646235 | 0.50 |
ENST00000556126.2
ENST00000579469.2 |
SMIM6
|
small integral membrane protein 6 |
| chr20_+_33217325 | 0.48 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr11_-_85686123 | 0.48 |
ENST00000316398.5
|
CCDC89
|
coiled-coil domain containing 89 |
| chr14_+_64914361 | 0.48 |
ENST00000607599.6
ENST00000548752.7 ENST00000551947.6 ENST00000549115.7 ENST00000552002.7 ENST00000551093.6 ENST00000549987.1 |
CHURC1
CHURC1-FNTB
|
churchill domain containing 1 CHURC1-FNTB readthrough |
| chr6_-_52763473 | 0.48 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr10_+_92834594 | 0.48 |
ENST00000371552.8
|
EXOC6
|
exocyst complex component 6 |
| chr1_+_236523417 | 0.47 |
ENST00000341872.10
ENST00000416919.6 ENST00000450372.6 |
LGALS8
|
galectin 8 |
| chr14_+_50533026 | 0.47 |
ENST00000441560.6
|
ATL1
|
atlastin GTPase 1 |
| chr3_+_69936583 | 0.47 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr2_+_161416273 | 0.47 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr6_-_119149124 | 0.46 |
ENST00000368475.8
|
FAM184A
|
family with sequence similarity 184 member A |
| chr20_+_13008919 | 0.46 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr18_-_55589795 | 0.46 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chrX_-_109482075 | 0.46 |
ENST00000218006.3
|
GUCY2F
|
guanylate cyclase 2F, retinal |
| chr16_+_69924984 | 0.45 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_-_41184895 | 0.45 |
ENST00000620667.1
ENST00000398472.2 |
KRTAP4-1
|
keratin associated protein 4-1 |
| chr19_+_44212525 | 0.45 |
ENST00000391961.6
ENST00000621083.4 ENST00000313040.12 ENST00000586228.5 ENST00000588219.5 ENST00000589707.5 ENST00000588394.5 ENST00000589005.5 |
ZNF227
|
zinc finger protein 227 |
| chr12_+_19205294 | 0.45 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr18_-_55589836 | 0.45 |
ENST00000537578.5
ENST00000564403.6 |
TCF4
|
transcription factor 4 |
| chr12_-_10130241 | 0.45 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr21_-_41926680 | 0.45 |
ENST00000329623.11
|
C2CD2
|
C2 calcium dependent domain containing 2 |
| chr10_+_99782628 | 0.44 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr12_+_109116581 | 0.44 |
ENST00000338432.11
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr15_-_55365231 | 0.44 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr5_+_73626158 | 0.44 |
ENST00000296794.10
ENST00000545377.5 ENST00000509848.5 ENST00000513042.7 |
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr18_+_13277351 | 0.43 |
ENST00000679091.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr9_-_120477354 | 0.42 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr7_-_64563063 | 0.42 |
ENST00000309683.11
|
ZNF680
|
zinc finger protein 680 |
| chr2_-_156342348 | 0.42 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr17_+_21126947 | 0.42 |
ENST00000579303.5
|
DHRS7B
|
dehydrogenase/reductase 7B |
| chr11_-_115504389 | 0.42 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr8_+_74339566 | 0.41 |
ENST00000675376.1
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr6_-_168079159 | 0.40 |
ENST00000283309.10
|
FRMD1
|
FERM domain containing 1 |
| chr10_-_48274567 | 0.40 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr8_+_109539696 | 0.40 |
ENST00000395785.6
|
EBAG9
|
estrogen receptor binding site associated antigen 9 |
| chr5_+_141245384 | 0.40 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chr12_-_49188811 | 0.40 |
ENST00000295766.9
|
TUBA1A
|
tubulin alpha 1a |
| chr5_-_20575850 | 0.39 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr12_-_10130143 | 0.39 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr1_+_174700413 | 0.39 |
ENST00000529145.6
ENST00000325589.9 |
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr4_+_41613476 | 0.39 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_57633161 | 0.39 |
ENST00000541801.5
ENST00000347302.7 ENST00000240731.5 ENST00000254182.11 ENST00000391703.3 |
ZNF211
|
zinc finger protein 211 |
| chr1_+_220748297 | 0.39 |
ENST00000366913.8
|
MTARC2
|
mitochondrial amidoxime reducing component 2 |
| chr9_-_13279407 | 0.38 |
ENST00000546205.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr10_+_91923762 | 0.38 |
ENST00000265990.11
|
BTAF1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr1_+_84164684 | 0.37 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr9_-_13279642 | 0.37 |
ENST00000319217.12
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr9_-_13279564 | 0.36 |
ENST00000541718.5
ENST00000447879.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr3_+_138621207 | 0.36 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr5_+_38403535 | 0.36 |
ENST00000336740.10
ENST00000397202.6 |
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr19_+_1077394 | 0.35 |
ENST00000590577.2
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr16_+_28751787 | 0.35 |
ENST00000357796.7
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family member B9 |
| chr2_+_162318884 | 0.35 |
ENST00000446271.5
ENST00000429691.6 |
GCA
|
grancalcin |
| chr2_-_216372432 | 0.34 |
ENST00000273067.5
|
MARCHF4
|
membrane associated ring-CH-type finger 4 |
| chr11_+_22666604 | 0.34 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr19_+_4198075 | 0.34 |
ENST00000262970.9
|
ANKRD24
|
ankyrin repeat domain 24 |
| chr5_-_218136 | 0.34 |
ENST00000296824.4
|
CCDC127
|
coiled-coil domain containing 127 |
| chr9_-_135961310 | 0.33 |
ENST00000371756.4
|
UBAC1
|
UBA domain containing 1 |
| chr5_-_111756245 | 0.33 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr5_+_141208697 | 0.33 |
ENST00000624949.1
ENST00000622978.1 ENST00000239450.4 |
PCDHB12
|
protocadherin beta 12 |
| chr13_-_99258366 | 0.32 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr9_+_75890664 | 0.32 |
ENST00000376767.7
ENST00000674117.1 ENST00000376752.8 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr14_+_101809795 | 0.32 |
ENST00000350249.7
ENST00000557621.5 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chr14_-_73950393 | 0.32 |
ENST00000651776.1
|
FAM161B
|
FAM161 centrosomal protein B |
| chr4_+_85475131 | 0.32 |
ENST00000395184.6
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_+_112176364 | 0.32 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr3_+_138621225 | 0.32 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_241532121 | 0.32 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chrX_+_109535775 | 0.32 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr4_+_145482761 | 0.31 |
ENST00000507367.1
ENST00000394092.6 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr1_+_204870831 | 0.31 |
ENST00000404076.5
ENST00000539706.6 |
NFASC
|
neurofascin |
| chr22_-_50245016 | 0.31 |
ENST00000248846.10
|
TUBGCP6
|
tubulin gamma complex associated protein 6 |
| chr17_-_41140487 | 0.31 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr6_-_168319691 | 0.31 |
ENST00000610183.1
ENST00000607983.1 |
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr1_+_42846573 | 0.31 |
ENST00000372502.5
ENST00000372508.7 ENST00000651192.1 ENST00000372507.5 ENST00000372506.5 ENST00000372504.5 |
ZNF691
|
zinc finger protein 691 |
| chr6_+_122789197 | 0.31 |
ENST00000368440.5
|
SMPDL3A
|
sphingomyelin phosphodiesterase acid like 3A |
| chr3_-_15341368 | 0.31 |
ENST00000408919.7
|
SH3BP5
|
SH3 domain binding protein 5 |
| chr11_-_107457801 | 0.30 |
ENST00000282251.9
|
CWF19L2
|
CWF19 like cell cycle control factor 2 |
| chr6_-_160258814 | 0.30 |
ENST00000366953.8
|
SLC22A2
|
solute carrier family 22 member 2 |
| chr11_+_58943249 | 0.30 |
ENST00000300079.9
|
GLYATL1
|
glycine-N-acyltransferase like 1 |
| chr5_-_24644968 | 0.30 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr19_-_7294406 | 0.29 |
ENST00000302850.10
|
INSR
|
insulin receptor |
| chr9_+_105662457 | 0.29 |
ENST00000334077.6
|
TAL2
|
TAL bHLH transcription factor 2 |
| chr6_-_168319762 | 0.28 |
ENST00000366795.4
|
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr5_+_141177790 | 0.28 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr7_-_32490361 | 0.28 |
ENST00000410044.5
ENST00000450169.7 ENST00000409987.5 ENST00000409782.5 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_+_173398438 | 0.28 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1 |
| chr20_-_17558811 | 0.28 |
ENST00000536626.7
ENST00000377868.6 |
BFSP1
|
beaded filament structural protein 1 |
| chr8_+_109539816 | 0.28 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated antigen 9 |
| chr6_-_116060859 | 0.27 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr1_+_231338242 | 0.27 |
ENST00000008440.9
ENST00000295050.12 |
SPRTN
|
SprT-like N-terminal domain |
| chr15_+_72654738 | 0.27 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family member B |
| chr14_+_22516273 | 0.27 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.8 | 2.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 1.8 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.3 | 1.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 1.0 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.3 | 2.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 4.0 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.2 | 0.6 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 3.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 5.3 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.6 | GO:1900108 | inner medullary collecting duct development(GO:0072061) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.4 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.4 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.8 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.5 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.9 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 1.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.6 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.1 | 0.7 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 6.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 0.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 2.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.6 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 4.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0002001 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) nerve growth factor processing(GO:0032455) |
| 0.1 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 3.1 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.8 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 1.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.5 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.6 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 2.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0072257 | contact inhibition(GO:0060242) metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0015866 | ADP transport(GO:0015866) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 0.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 2.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.8 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 5.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 3.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.0 | 5.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.5 | 3.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 3.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 0.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 1.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 2.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 5.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.8 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.3 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.2 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.1 | 1.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 5.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.9 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 3.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.0 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 5.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 4.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 5.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |