Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for GATA5
Z-value: 0.42
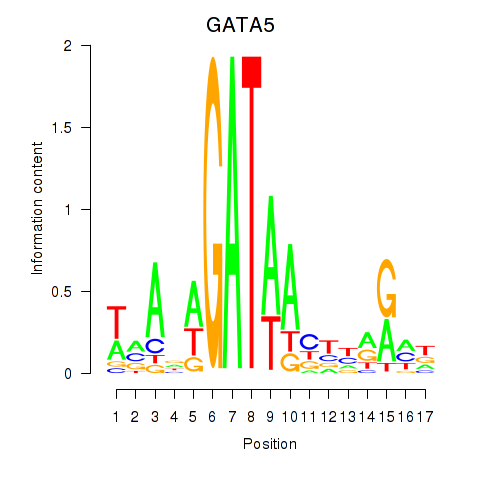
Transcription factors associated with GATA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA5
|
ENSG00000130700.7 | GATA5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA5 | hg38_v1_chr20_-_62475983_62476001 | 0.18 | 3.5e-01 | Click! |
Activity profile of GATA5 motif
Sorted Z-values of GATA5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_4786354 | 0.80 |
ENST00000328739.6
ENST00000441199.2 ENST00000416307.6 |
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr4_+_73740541 | 0.68 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr17_-_4786433 | 0.57 |
ENST00000354194.4
|
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr4_+_72031902 | 0.54 |
ENST00000344413.6
ENST00000308744.12 |
NPFFR2
|
neuropeptide FF receptor 2 |
| chr6_+_85449584 | 0.49 |
ENST00000369651.7
|
NT5E
|
5'-nucleotidase ecto |
| chr6_+_42050876 | 0.48 |
ENST00000465926.5
ENST00000482432.1 |
TAF8
|
TATA-box binding protein associated factor 8 |
| chr17_+_64449037 | 0.43 |
ENST00000615220.4
ENST00000619286.5 ENST00000612535.4 ENST00000616498.4 |
MILR1
|
mast cell immunoglobulin like receptor 1 |
| chr4_-_109801978 | 0.38 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr7_+_142760398 | 0.37 |
ENST00000632998.1
|
PRSS2
|
serine protease 2 |
| chr2_+_190927649 | 0.37 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr2_+_102311502 | 0.35 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr2_+_101998955 | 0.34 |
ENST00000393414.6
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr1_+_202462730 | 0.31 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chrX_+_105948429 | 0.30 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr12_+_53097656 | 0.28 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr1_+_21570303 | 0.28 |
ENST00000374830.2
|
ALPL
|
alkaline phosphatase, biomineralization associated |
| chr18_+_63775395 | 0.25 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr15_+_40953463 | 0.24 |
ENST00000617768.5
|
CHAC1
|
ChaC glutathione specific gamma-glutamylcyclotransferase 1 |
| chr8_-_23854796 | 0.24 |
ENST00000290271.7
|
STC1
|
stanniocalcin 1 |
| chr4_-_99657820 | 0.20 |
ENST00000511828.2
|
C4orf54
|
chromosome 4 open reading frame 54 |
| chr3_-_138132381 | 0.19 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr4_+_109912877 | 0.18 |
ENST00000265171.10
ENST00000509793.5 ENST00000652245.1 |
EGF
|
epidermal growth factor |
| chr7_+_142749465 | 0.18 |
ENST00000486171.5
ENST00000619214.4 ENST00000311737.12 |
PRSS1
|
serine protease 1 |
| chr9_-_35650902 | 0.18 |
ENST00000259608.8
ENST00000618781.1 |
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr17_-_41047267 | 0.18 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chrX_+_153179276 | 0.17 |
ENST00000356661.7
|
MAGEA1
|
MAGE family member A1 |
| chr1_-_158686700 | 0.17 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1 |
| chr10_+_7703340 | 0.17 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr8_+_40153475 | 0.17 |
ENST00000315792.5
|
TCIM
|
transcriptional and immune response regulator |
| chr6_+_29396555 | 0.16 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr6_+_18387326 | 0.15 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B |
| chr1_+_56854764 | 0.15 |
ENST00000361249.4
|
C8A
|
complement C8 alpha chain |
| chr12_-_30735014 | 0.15 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr7_-_143647646 | 0.15 |
ENST00000636941.1
|
TCAF2C
|
TRPM8 channel associated factor 2C |
| chr8_-_90082871 | 0.15 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr12_+_20815672 | 0.15 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr12_-_14814160 | 0.15 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr8_+_12108172 | 0.15 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D |
| chr2_+_96642729 | 0.14 |
ENST00000624922.6
ENST00000623019.5 |
FER1L5
|
fer-1 like family member 5 |
| chr12_-_8650529 | 0.14 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr11_+_30231000 | 0.14 |
ENST00000254122.8
ENST00000533718.2 ENST00000417547.1 |
FSHB
|
follicle stimulating hormone subunit beta |
| chr10_+_102152169 | 0.14 |
ENST00000405356.5
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr1_+_24319342 | 0.13 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr13_+_30735523 | 0.13 |
ENST00000380490.5
|
ALOX5AP
|
arachidonate 5-lipoxygenase activating protein |
| chr7_-_123557679 | 0.13 |
ENST00000677264.1
ENST00000471770.5 ENST00000676730.1 ENST00000677391.1 ENST00000677251.1 ENST00000470123.2 ENST00000679163.1 ENST00000676614.1 ENST00000378795.9 ENST00000676516.1 ENST00000355749.7 ENST00000678090.1 |
NDUFA5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr1_+_206635703 | 0.13 |
ENST00000649163.1
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr11_+_56176618 | 0.13 |
ENST00000312298.1
|
OR5J2
|
olfactory receptor family 5 subfamily J member 2 |
| chr20_+_45629717 | 0.13 |
ENST00000372643.4
|
WFDC10A
|
WAP four-disulfide core domain 10A |
| chr2_-_46916020 | 0.13 |
ENST00000409800.5
ENST00000409218.5 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr1_-_153041111 | 0.13 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr1_+_206635573 | 0.12 |
ENST00000367108.7
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr8_-_6999198 | 0.12 |
ENST00000382689.8
|
DEFA1B
|
defensin alpha 1B |
| chr20_-_45705019 | 0.12 |
ENST00000330523.9
ENST00000335769.2 |
WFDC10B
|
WAP four-disulfide core domain 10B |
| chr17_+_47522931 | 0.12 |
ENST00000525007.5
ENST00000530173.6 |
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr2_+_29113989 | 0.11 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr20_-_31406207 | 0.11 |
ENST00000376314.3
|
DEFB121
|
defensin beta 121 |
| chr4_+_154563003 | 0.10 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr4_-_64409444 | 0.10 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr19_-_41844629 | 0.10 |
ENST00000609812.6
|
LYPD4
|
LY6/PLAUR domain containing 4 |
| chr2_+_201132928 | 0.10 |
ENST00000462763.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr5_-_148654522 | 0.10 |
ENST00000377888.8
|
HTR4
|
5-hydroxytryptamine receptor 4 |
| chr13_-_102759059 | 0.10 |
ENST00000322527.4
|
CCDC168
|
coiled-coil domain containing 168 |
| chr3_+_184362599 | 0.10 |
ENST00000455712.5
|
POLR2H
|
RNA polymerase II, I and III subunit H |
| chr1_+_103617427 | 0.10 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr19_+_926001 | 0.10 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr9_+_72628020 | 0.10 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr2_+_201132958 | 0.10 |
ENST00000479953.6
ENST00000340870.6 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr4_-_163332589 | 0.09 |
ENST00000296533.3
ENST00000509586.5 ENST00000504391.5 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr4_+_70195719 | 0.09 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated |
| chr5_+_93583212 | 0.09 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr17_-_59707404 | 0.09 |
ENST00000393038.3
|
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr8_-_7018295 | 0.09 |
ENST00000327857.7
|
DEFA3
|
defensin alpha 3 |
| chr11_-_101129806 | 0.09 |
ENST00000325455.10
ENST00000617858.4 ENST00000619228.2 |
PGR
|
progesterone receptor |
| chr2_-_108989206 | 0.08 |
ENST00000258443.7
ENST00000409271.5 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr15_+_58138368 | 0.08 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr2_+_201132769 | 0.08 |
ENST00000494258.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr6_-_132757883 | 0.08 |
ENST00000525289.5
ENST00000326499.11 |
VNN2
|
vanin 2 |
| chr14_+_21768482 | 0.08 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chrM_+_12329 | 0.08 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr2_-_105396943 | 0.08 |
ENST00000409807.5
|
FHL2
|
four and a half LIM domains 2 |
| chr2_+_201132872 | 0.08 |
ENST00000470178.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr6_-_169250825 | 0.08 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr1_+_196774813 | 0.08 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr1_+_86468902 | 0.08 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr6_-_134950081 | 0.08 |
ENST00000367847.2
ENST00000265605.7 ENST00000367845.6 |
ALDH8A1
|
aldehyde dehydrogenase 8 family member A1 |
| chr14_-_106154113 | 0.07 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr6_+_29395631 | 0.07 |
ENST00000642051.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr9_-_112175185 | 0.07 |
ENST00000355396.7
|
SUSD1
|
sushi domain containing 1 |
| chr9_-_90642791 | 0.07 |
ENST00000375765.5
ENST00000636786.1 |
DIRAS2
|
DIRAS family GTPase 2 |
| chr14_-_106538331 | 0.07 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr1_+_23743153 | 0.07 |
ENST00000418390.6
|
ELOA
|
elongin A |
| chr9_-_41189310 | 0.07 |
ENST00000456520.5
ENST00000377391.8 ENST00000613716.4 ENST00000617933.1 |
CBWD6
|
COBW domain containing 6 |
| chr7_+_136868622 | 0.07 |
ENST00000680005.1
ENST00000445907.6 |
CHRM2
|
cholinergic receptor muscarinic 2 |
| chr15_-_63381835 | 0.07 |
ENST00000344366.7
ENST00000178638.8 ENST00000422263.2 |
CA12
|
carbonic anhydrase 12 |
| chr18_+_58362467 | 0.07 |
ENST00000675101.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr13_-_46182136 | 0.07 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr6_-_89819699 | 0.06 |
ENST00000439638.1
ENST00000629399.2 ENST00000369393.8 |
MDN1
|
midasin AAA ATPase 1 |
| chr3_+_141426108 | 0.06 |
ENST00000441582.2
ENST00000510726.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chrX_-_146814726 | 0.06 |
ENST00000458472.1
|
CXorf51A
|
chromosome X open reading frame 51A |
| chrX_+_146809768 | 0.06 |
ENST00000438525.3
|
CXorf51B
|
chromosome X open reading frame 51B |
| chr3_+_172039556 | 0.06 |
ENST00000415807.7
ENST00000421757.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr12_-_68225806 | 0.06 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr11_+_114439424 | 0.06 |
ENST00000544196.5
ENST00000265881.10 ENST00000539754.5 ENST00000539275.5 |
REXO2
|
RNA exonuclease 2 |
| chr9_+_104094557 | 0.06 |
ENST00000374787.7
|
SMC2
|
structural maintenance of chromosomes 2 |
| chr9_+_68241854 | 0.06 |
ENST00000616550.4
ENST00000618217.4 ENST00000377342.9 ENST00000478048.5 ENST00000360171.11 |
CBWD3
|
COBW domain containing 3 |
| chr5_+_132060648 | 0.06 |
ENST00000296870.3
|
IL3
|
interleukin 3 |
| chr6_+_146029059 | 0.06 |
ENST00000282753.6
|
GRM1
|
glutamate metabotropic receptor 1 |
| chr3_+_178558700 | 0.06 |
ENST00000432997.5
ENST00000455865.5 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr17_-_47831509 | 0.06 |
ENST00000414011.1
ENST00000351111.7 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr2_-_182866627 | 0.06 |
ENST00000295113.5
|
FRZB
|
frizzled related protein |
| chr9_-_112175264 | 0.05 |
ENST00000374264.6
|
SUSD1
|
sushi domain containing 1 |
| chrX_+_30243715 | 0.05 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr4_+_141636923 | 0.05 |
ENST00000529613.5
|
IL15
|
interleukin 15 |
| chr7_+_142791635 | 0.05 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr4_-_46389351 | 0.05 |
ENST00000503806.5
ENST00000356504.5 ENST00000514090.5 ENST00000506961.5 |
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr6_-_154356735 | 0.05 |
ENST00000367220.8
ENST00000265198.8 ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_156142962 | 0.05 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr20_-_50958520 | 0.05 |
ENST00000371588.10
ENST00000371584.9 ENST00000413082.1 ENST00000371582.8 |
DPM1
|
dolichyl-phosphate mannosyltransferase subunit 1, catalytic |
| chr9_-_90642855 | 0.05 |
ENST00000637905.1
|
DIRAS2
|
DIRAS family GTPase 2 |
| chr14_+_66824439 | 0.05 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr6_-_137044269 | 0.05 |
ENST00000635289.1
ENST00000541547.5 |
IL20RA
|
interleukin 20 receptor subunit alpha |
| chr3_+_98147479 | 0.05 |
ENST00000641380.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr13_-_108218293 | 0.05 |
ENST00000405925.2
|
LIG4
|
DNA ligase 4 |
| chr11_-_61967127 | 0.05 |
ENST00000532601.1
|
FTH1
|
ferritin heavy chain 1 |
| chr10_+_5048748 | 0.05 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr8_+_117135259 | 0.05 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr5_+_38445539 | 0.05 |
ENST00000397210.7
ENST00000506135.5 ENST00000508131.5 |
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr12_+_101594849 | 0.04 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr11_+_124241095 | 0.04 |
ENST00000641972.1
|
OR8G1
|
olfactory receptor family 8 subfamily G member 1 |
| chr21_-_30480364 | 0.04 |
ENST00000390689.3
|
KRTAP19-1
|
keratin associated protein 19-1 |
| chr8_-_78805515 | 0.04 |
ENST00000379113.6
ENST00000541183.2 |
IL7
|
interleukin 7 |
| chr8_-_78805306 | 0.04 |
ENST00000639719.1
ENST00000263851.9 |
IL7
|
interleukin 7 |
| chr8_-_140800535 | 0.04 |
ENST00000521986.5
ENST00000523539.5 |
PTK2
|
protein tyrosine kinase 2 |
| chrX_-_77895546 | 0.04 |
ENST00000358075.11
|
MAGT1
|
magnesium transporter 1 |
| chr10_+_18400562 | 0.04 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr16_+_33827140 | 0.04 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr3_-_108757407 | 0.04 |
ENST00000295755.7
|
RETNLB
|
resistin like beta |
| chr4_+_158210479 | 0.04 |
ENST00000504569.5
ENST00000509278.5 ENST00000514558.5 ENST00000503200.5 ENST00000296529.11 |
TMEM144
|
transmembrane protein 144 |
| chr3_+_108822759 | 0.04 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_+_248095184 | 0.04 |
ENST00000358120.3
ENST00000641893.1 |
OR2L13
|
olfactory receptor family 2 subfamily L member 13 |
| chr3_-_185923893 | 0.04 |
ENST00000259043.11
|
TRA2B
|
transformer 2 beta homolog |
| chr3_+_46242453 | 0.04 |
ENST00000452454.1
ENST00000395940.3 ENST00000457243.1 |
CCR3
|
C-C motif chemokine receptor 3 |
| chr6_-_159044980 | 0.04 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr3_+_108822778 | 0.03 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr3_-_108058361 | 0.03 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr11_+_62270150 | 0.03 |
ENST00000525380.1
ENST00000227918.3 |
SCGB2A2
|
secretoglobin family 2A member 2 |
| chr3_+_44874606 | 0.03 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr19_-_44304968 | 0.03 |
ENST00000591609.1
ENST00000589799.5 ENST00000291182.9 ENST00000650576.1 ENST00000589248.5 |
ZNF235
|
zinc finger protein 235 |
| chr4_+_70592253 | 0.03 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr1_-_44141631 | 0.03 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr6_+_152697888 | 0.03 |
ENST00000367245.5
ENST00000529453.1 |
MYCT1
|
MYC target 1 |
| chr10_+_88664439 | 0.03 |
ENST00000394375.7
ENST00000608620.5 ENST00000238983.9 ENST00000355843.2 |
LIPF
|
lipase F, gastric type |
| chr3_-_179251615 | 0.03 |
ENST00000314235.9
ENST00000392685.6 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr13_+_31739520 | 0.03 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr16_+_56451513 | 0.03 |
ENST00000562150.5
ENST00000561646.5 ENST00000566157.6 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron dependent oxygenase domain containing 1 |
| chr14_+_61697622 | 0.03 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr6_+_29461440 | 0.03 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr15_+_76336755 | 0.03 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chrX_+_11759604 | 0.03 |
ENST00000649602.1
ENST00000650215.1 ENST00000649685.1 ENST00000649649.1 ENST00000380693.8 ENST00000380692.7 ENST00000650628.1 ENST00000649851.1 ENST00000421368.3 |
MSL3
|
MSL complex subunit 3 |
| chr12_+_71686473 | 0.03 |
ENST00000549735.5
|
TMEM19
|
transmembrane protein 19 |
| chr9_-_6645712 | 0.03 |
ENST00000321612.8
|
GLDC
|
glycine decarboxylase |
| chr10_+_94402486 | 0.03 |
ENST00000225235.5
|
TBC1D12
|
TBC1 domain family member 12 |
| chr3_-_183099508 | 0.03 |
ENST00000476176.5
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 |
| chr20_-_25623945 | 0.03 |
ENST00000304788.4
|
NANP
|
N-acetylneuraminic acid phosphatase |
| chr12_+_57488059 | 0.03 |
ENST00000628866.2
ENST00000262027.10 |
MARS1
|
methionyl-tRNA synthetase 1 |
| chr3_+_46242371 | 0.02 |
ENST00000545097.1
|
CCR3
|
C-C motif chemokine receptor 3 |
| chr1_+_162381703 | 0.02 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr4_+_145625992 | 0.02 |
ENST00000541599.5
|
MMAA
|
metabolism of cobalamin associated A |
| chr17_-_15598797 | 0.02 |
ENST00000354433.7
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr7_-_87059639 | 0.02 |
ENST00000450689.7
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr16_-_69339493 | 0.02 |
ENST00000562595.5
ENST00000615447.1 ENST00000306875.10 ENST00000562081.2 |
COG8
|
component of oligomeric golgi complex 8 |
| chr19_-_36489854 | 0.02 |
ENST00000493391.6
|
ZNF566
|
zinc finger protein 566 |
| chr15_+_84237586 | 0.02 |
ENST00000512109.1
|
GOLGA6L4
|
golgin A6 family like 4 |
| chr9_+_2110354 | 0.02 |
ENST00000634772.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_181985846 | 0.02 |
ENST00000682840.1
ENST00000409137.7 ENST00000280295.7 |
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C |
| chr5_+_145936554 | 0.02 |
ENST00000359120.9
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr11_-_26722051 | 0.02 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr6_+_152697934 | 0.02 |
ENST00000532295.1
|
MYCT1
|
MYC target 1 |
| chr18_+_13612614 | 0.02 |
ENST00000586765.1
ENST00000677910.1 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr9_+_89318492 | 0.02 |
ENST00000375807.8
ENST00000339901.8 |
SECISBP2
|
SECIS binding protein 2 |
| chr14_+_20455968 | 0.02 |
ENST00000438886.1
|
APEX1
|
apurinic/apyrimidinic endodeoxyribonuclease 1 |
| chr19_+_35329161 | 0.02 |
ENST00000341773.10
ENST00000600131.5 ENST00000595780.5 ENST00000597916.5 ENST00000593867.5 ENST00000600424.5 ENST00000599811.5 ENST00000536635.6 ENST00000085219.10 ENST00000544992.6 ENST00000419549.6 |
CD22
|
CD22 molecule |
| chrX_-_77895414 | 0.02 |
ENST00000618282.5
ENST00000373336.3 |
MAGT1
|
magnesium transporter 1 |
| chr12_-_70637405 | 0.02 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr6_-_130956371 | 0.02 |
ENST00000639623.1
ENST00000525193.5 ENST00000527659.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr1_+_75786246 | 0.02 |
ENST00000319942.8
|
RABGGTB
|
Rab geranylgeranyltransferase subunit beta |
| chr13_+_31739542 | 0.02 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr3_-_143848442 | 0.02 |
ENST00000474151.1
ENST00000316549.11 |
SLC9A9
|
solute carrier family 9 member A9 |
| chr19_-_36489435 | 0.02 |
ENST00000434377.6
ENST00000424129.7 ENST00000452939.6 ENST00000427002.2 |
ZNF566
|
zinc finger protein 566 |
| chr19_-_36489553 | 0.02 |
ENST00000392170.7
|
ZNF566
|
zinc finger protein 566 |
| chr5_-_159099684 | 0.02 |
ENST00000380654.8
|
EBF1
|
EBF transcription factor 1 |
| chr6_+_118548289 | 0.02 |
ENST00000357525.6
|
PLN
|
phospholamban |
| chr18_+_13611764 | 0.01 |
ENST00000585931.5
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr5_-_159099745 | 0.01 |
ENST00000517373.1
|
EBF1
|
EBF transcription factor 1 |
| chr6_-_170553216 | 0.01 |
ENST00000262193.7
|
PSMB1
|
proteasome 20S subunit beta 1 |
| chrM_+_10055 | 0.01 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chr10_+_7703300 | 0.01 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_-_82844418 | 0.01 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr11_-_85665077 | 0.01 |
ENST00000527447.2
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr15_+_88635626 | 0.01 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.3 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.5 | GO:0046086 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.1 | 0.7 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.4 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0015855 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0060011 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.0 | GO:0016487 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.5 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.0 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |