|
chr2_+_102311502
Show fit
|
3.15 |
ENST00000404917.6
ENST00000410040.5
|
IL1RL1
IL18R1
|
interleukin 1 receptor like 1
interleukin 18 receptor 1
|
|
chr7_+_142760398
Show fit
|
1.62 |
ENST00000632998.1
|
PRSS2
|
serine protease 2
|
|
chr9_+_33795551
Show fit
|
1.36 |
ENST00000379405.4
|
PRSS3
|
serine protease 3
|
|
chr17_+_76385256
Show fit
|
1.17 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1
|
|
chr20_-_56392131
Show fit
|
1.00 |
ENST00000422322.5
ENST00000371356.6
ENST00000451915.1
ENST00000347343.6
ENST00000395911.5
ENST00000395915.8
ENST00000395907.5
ENST00000441357.5
ENST00000456249.5
ENST00000420474.5
ENST00000395914.5
ENST00000312783.10
ENST00000395913.7
|
AURKA
|
aurora kinase A
|
|
chr7_+_155298561
Show fit
|
0.86 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1
|
|
chr6_+_12290353
Show fit
|
0.76 |
ENST00000379375.6
|
EDN1
|
endothelin 1
|
|
chr7_+_142749465
Show fit
|
0.74 |
ENST00000486171.5
ENST00000619214.4
ENST00000311737.12
|
PRSS1
|
serine protease 1
|
|
chr4_-_102345196
Show fit
|
0.74 |
ENST00000683412.1
ENST00000682227.1
|
SLC39A8
|
solute carrier family 39 member 8
|
|
chr1_-_24143112
Show fit
|
0.73 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1
|
|
chrX_+_38006551
Show fit
|
0.73 |
ENST00000297875.7
|
SYTL5
|
synaptotagmin like 5
|
|
chr8_+_123182635
Show fit
|
0.67 |
ENST00000276699.10
ENST00000522648.5
|
FAM83A
|
family with sequence similarity 83 member A
|
|
chr1_-_93585071
Show fit
|
0.65 |
ENST00000539242.5
|
BCAR3
|
BCAR3 adaptor protein, NSP family member
|
|
chr12_+_4321197
Show fit
|
0.62 |
ENST00000179259.6
|
TIGAR
|
TP53 induced glycolysis regulatory phosphatase
|
|
chr2_-_68871382
Show fit
|
0.56 |
ENST00000295379.2
|
BMP10
|
bone morphogenetic protein 10
|
|
chr9_-_112175264
Show fit
|
0.55 |
ENST00000374264.6
|
SUSD1
|
sushi domain containing 1
|
|
chr9_-_112175185
Show fit
|
0.51 |
ENST00000355396.7
|
SUSD1
|
sushi domain containing 1
|
|
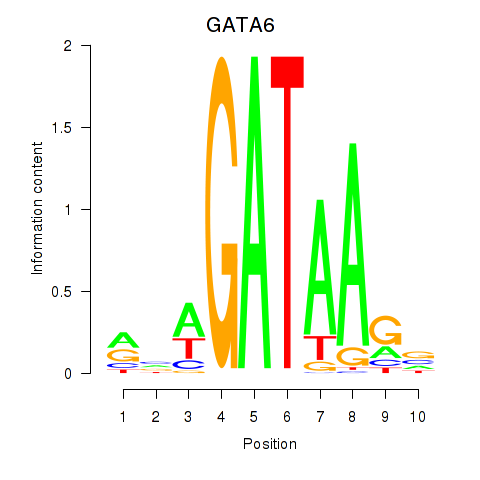
chr18_+_22169580
Show fit
|
0.49 |
ENST00000269216.10
|
GATA6
|
GATA binding protein 6
|
|
chr5_+_132073782
Show fit
|
0.46 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2
|
|
chrX_+_108044967
Show fit
|
0.41 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr19_+_10286971
Show fit
|
0.40 |
ENST00000340992.4
ENST00000393717.2
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|
|
chrX_+_108045050
Show fit
|
0.40 |
ENST00000458383.1
ENST00000217957.10
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr19_+_10286944
Show fit
|
0.39 |
ENST00000380770.5
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|
|
chr2_-_43838832
Show fit
|
0.38 |
ENST00000405322.8
|
ABCG5
|
ATP binding cassette subfamily G member 5
|
|
chr9_-_112175283
Show fit
|
0.37 |
ENST00000374270.8
ENST00000374263.7
|
SUSD1
|
sushi domain containing 1
|
|
chr10_-_127892930
Show fit
|
0.37 |
ENST00000368671.4
|
CLRN3
|
clarin 3
|
|
chr1_-_158686700
Show fit
|
0.33 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1
|
|
chr8_+_69492793
Show fit
|
0.33 |
ENST00000616868.1
ENST00000419716.7
ENST00000402687.9
|
SULF1
|
sulfatase 1
|
|
chr11_-_66438788
Show fit
|
0.32 |
ENST00000329819.4
ENST00000310999.11
ENST00000430466.6
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr1_-_225999312
Show fit
|
0.31 |
ENST00000272091.8
|
SDE2
|
SDE2 telomere maintenance homolog
|
|
chr17_+_19411220
Show fit
|
0.31 |
ENST00000461366.2
|
RNF112
|
ring finger protein 112
|
|
chr13_-_46142834
Show fit
|
0.30 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1
|
|
chr4_-_102345061
Show fit
|
0.28 |
ENST00000394833.6
|
SLC39A8
|
solute carrier family 39 member 8
|
|
chr19_-_38812936
Show fit
|
0.26 |
ENST00000307751.9
ENST00000594209.1
|
LGALS4
|
galectin 4
|
|
chr17_+_77287891
Show fit
|
0.25 |
ENST00000449803.6
ENST00000431235.6
|
SEPTIN9
|
septin 9
|
|
chr6_+_130018565
Show fit
|
0.22 |
ENST00000361794.7
ENST00000526087.5
ENST00000533560.5
|
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3
|
|
chr21_-_38498415
Show fit
|
0.22 |
ENST00000398905.5
ENST00000398907.5
ENST00000453032.6
ENST00000288319.12
|
ERG
|
ETS transcription factor ERG
|
|
chr1_-_167518521
Show fit
|
0.22 |
ENST00000362089.10
|
CD247
|
CD247 molecule
|
|
chr1_-_167518583
Show fit
|
0.21 |
ENST00000392122.3
|
CD247
|
CD247 molecule
|
|
chr8_-_94436926
Show fit
|
0.21 |
ENST00000481490.3
|
FSBP
|
fibrinogen silencer binding protein
|
|
chr2_+_102311546
Show fit
|
0.21 |
ENST00000233954.6
ENST00000447231.5
|
IL1RL1
|
interleukin 1 receptor like 1
|
|
chr8_+_21966215
Show fit
|
0.21 |
ENST00000433566.8
|
XPO7
|
exportin 7
|
|
chr7_-_142962206
Show fit
|
0.20 |
ENST00000460479.2
ENST00000476829.5
ENST00000355265.7
|
KEL
|
Kell metallo-endopeptidase (Kell blood group)
|
|
chr11_+_10305065
Show fit
|
0.20 |
ENST00000534464.1
ENST00000278175.10
ENST00000530439.1
ENST00000524948.5
ENST00000528655.5
ENST00000526492.4
ENST00000525063.2
|
ADM
|
adrenomedullin
|
|
chr18_-_31162849
Show fit
|
0.17 |
ENST00000257197.7
ENST00000257198.6
|
DSC1
|
desmocollin 1
|
|
chr10_+_69315760
Show fit
|
0.15 |
ENST00000298649.8
|
HK1
|
hexokinase 1
|
|
chr8_-_41797589
Show fit
|
0.14 |
ENST00000347528.8
ENST00000289734.13
|
ANK1
|
ankyrin 1
|
|
chr6_-_30160880
Show fit
|
0.14 |
ENST00000376704.3
|
TRIM10
|
tripartite motif containing 10
|
|
chr10_+_116590956
Show fit
|
0.13 |
ENST00000358834.9
ENST00000528052.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chr1_-_206023889
Show fit
|
0.13 |
ENST00000358184.7
ENST00000360218.3
ENST00000678712.1
ENST00000678498.1
|
CTSE
|
cathepsin E
|
|
chr1_+_66534171
Show fit
|
0.12 |
ENST00000682762.1
ENST00000424320.6
|
SGIP1
|
SH3GL interacting endocytic adaptor 1
|
|
chr1_-_42164737
Show fit
|
0.12 |
ENST00000357001.3
|
GUCA2A
|
guanylate cyclase activator 2A
|
|
chr10_+_116591010
Show fit
|
0.12 |
ENST00000530319.5
ENST00000527980.5
ENST00000471549.5
ENST00000534537.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chr6_+_31706866
Show fit
|
0.11 |
ENST00000375832.5
ENST00000503322.1
|
LY6G6F
LY6G6F-LY6G6D
|
lymphocyte antigen 6 family member G6F
LY6G6F-LY6G6D readthrough
|
|
chr22_-_29388530
Show fit
|
0.11 |
ENST00000357586.7
ENST00000432560.6
ENST00000405198.6
ENST00000317368.11
|
AP1B1
|
adaptor related protein complex 1 subunit beta 1
|
|
chr19_-_13102848
Show fit
|
0.10 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member
|
|
chr19_+_14028148
Show fit
|
0.10 |
ENST00000431365.3
ENST00000585987.1
|
RLN3
|
relaxin 3
|
|
chr2_+_219572304
Show fit
|
0.10 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha
|
|
chr4_+_15374541
Show fit
|
0.09 |
ENST00000382383.7
ENST00000429690.5
|
C1QTNF7
|
C1q and TNF related 7
|
|
chr6_-_30161200
Show fit
|
0.09 |
ENST00000449742.7
|
TRIM10
|
tripartite motif containing 10
|
|
chr19_-_51019699
Show fit
|
0.09 |
ENST00000358789.8
|
KLK10
|
kallikrein related peptidase 10
|
|
chr11_-_57390636
Show fit
|
0.09 |
ENST00000525955.1
ENST00000533605.5
ENST00000311862.10
|
PRG2
|
proteoglycan 2, pro eosinophil major basic protein
|
|
chr17_+_4932285
Show fit
|
0.09 |
ENST00000611961.1
|
GP1BA
|
glycoprotein Ib platelet subunit alpha
|
|
chr17_-_40782544
Show fit
|
0.08 |
ENST00000301656.4
|
KRT27
|
keratin 27
|
|
chr9_-_114930508
Show fit
|
0.08 |
ENST00000223795.3
ENST00000618336.4
|
TNFSF8
|
TNF superfamily member 8
|
|
chr5_+_1201588
Show fit
|
0.08 |
ENST00000304460.11
|
SLC6A19
|
solute carrier family 6 member 19
|
|
chr8_+_69466617
Show fit
|
0.08 |
ENST00000525061.5
ENST00000260128.8
ENST00000458141.6
|
SULF1
|
sulfatase 1
|
|
chr5_-_135954962
Show fit
|
0.08 |
ENST00000522943.5
ENST00000514447.2
ENST00000274507.6
|
LECT2
|
leukocyte cell derived chemotaxin 2
|
|
chr1_+_25272527
Show fit
|
0.08 |
ENST00000342055.9
ENST00000357542.8
ENST00000417538.6
ENST00000423810.6
ENST00000568195.5
|
RHD
|
Rh blood group D antigen
|
|
chr7_-_100641507
Show fit
|
0.08 |
ENST00000431692.5
ENST00000223051.8
|
TFR2
|
transferrin receptor 2
|
|
chr5_-_33984636
Show fit
|
0.07 |
ENST00000382102.7
ENST00000509381.1
|
SLC45A2
|
solute carrier family 45 member 2
|
|
chr12_+_69825221
Show fit
|
0.07 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like
|
|
chr19_+_35358460
Show fit
|
0.07 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3
|
|
chr1_+_25272492
Show fit
|
0.06 |
ENST00000454452.6
|
RHD
|
Rh blood group D antigen
|
|
chr18_+_45724127
Show fit
|
0.06 |
ENST00000619403.4
ENST00000587601.5
|
SLC14A1
|
solute carrier family 14 member 1 (Kidd blood group)
|
|
chr1_+_50108856
Show fit
|
0.06 |
ENST00000650764.1
ENST00000494555.2
ENST00000371824.7
ENST00000371823.8
ENST00000652693.1
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr12_-_55827546
Show fit
|
0.06 |
ENST00000546837.5
|
ENSG00000257390.5
|
novel protein
|
|
chr19_+_35358821
Show fit
|
0.06 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3
|
|
chr17_+_4932248
Show fit
|
0.06 |
ENST00000329125.6
|
GP1BA
|
glycoprotein Ib platelet subunit alpha
|
|
chr4_+_99574812
Show fit
|
0.06 |
ENST00000422897.6
ENST00000265517.10
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr19_-_11384299
Show fit
|
0.06 |
ENST00000592375.6
ENST00000222139.11
|
EPOR
|
erythropoietin receptor
|
|
chr20_+_44401269
Show fit
|
0.05 |
ENST00000443598.6
ENST00000415691.2
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr4_-_119322128
Show fit
|
0.05 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2
|
|
chr8_-_144792380
Show fit
|
0.05 |
ENST00000532702.1
ENST00000394920.6
ENST00000528957.6
ENST00000527914.5
|
RPL8
|
ribosomal protein L8
|
|
chr11_-_59845496
Show fit
|
0.05 |
ENST00000257248.3
|
CBLIF
|
cobalamin binding intrinsic factor
|
|
chr12_+_69825273
Show fit
|
0.05 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like
|
|
chr17_-_64006880
Show fit
|
0.05 |
ENST00000449662.6
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr20_-_32207708
Show fit
|
0.05 |
ENST00000246229.5
|
PLAGL2
|
PLAG1 like zinc finger 2
|
|
chr3_+_152268920
Show fit
|
0.05 |
ENST00000495875.6
ENST00000324210.10
ENST00000493459.5
|
MBNL1
|
muscleblind like splicing regulator 1
|
|
chr7_+_128739292
Show fit
|
0.05 |
ENST00000535011.6
ENST00000542996.6
ENST00000249364.9
ENST00000449187.6
|
CALU
|
calumenin
|
|
chr20_+_45306834
Show fit
|
0.05 |
ENST00000343694.8
ENST00000372741.7
ENST00000372743.5
|
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region like
|
|
chr3_+_148865288
Show fit
|
0.05 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3
|
|
chr6_-_46080332
Show fit
|
0.04 |
ENST00000185206.12
|
CLIC5
|
chloride intracellular channel 5
|
|
chr7_+_128739395
Show fit
|
0.04 |
ENST00000479257.5
|
CALU
|
calumenin
|
|
chr5_+_136160986
Show fit
|
0.04 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5
|
|
chr2_+_172928165
Show fit
|
0.04 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr18_+_45724172
Show fit
|
0.04 |
ENST00000321925.9
ENST00000415427.7
ENST00000589322.7
ENST00000586951.6
ENST00000535474.5
ENST00000402943.6
|
SLC14A1
|
solute carrier family 14 member 1 (Kidd blood group)
|
|
chr11_+_119087979
Show fit
|
0.04 |
ENST00000535253.5
ENST00000392841.1
ENST00000648374.1
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr20_+_56392607
Show fit
|
0.04 |
ENST00000217109.9
ENST00000452950.1
|
CSTF1
|
cleavage stimulation factor subunit 1
|
|
chr5_-_33984681
Show fit
|
0.04 |
ENST00000296589.9
|
SLC45A2
|
solute carrier family 45 member 2
|
|
chr14_+_21477177
Show fit
|
0.03 |
ENST00000448790.7
ENST00000673643.1
ENST00000457430.2
ENST00000673911.1
|
TOX4
|
TOX high mobility group box family member 4
|
|
chr1_+_154321107
Show fit
|
0.03 |
ENST00000484864.1
|
AQP10
|
aquaporin 10
|
|
chr1_+_25272479
Show fit
|
0.02 |
ENST00000622561.4
|
RHD
|
Rh blood group D antigen
|
|
chr17_+_58755821
Show fit
|
0.02 |
ENST00000308249.4
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent 1E
|
|
chr18_+_3447562
Show fit
|
0.02 |
ENST00000618001.4
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr15_+_49423233
Show fit
|
0.02 |
ENST00000560270.1
ENST00000267843.9
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7
|
|
chr15_+_58987652
Show fit
|
0.02 |
ENST00000348370.9
ENST00000559160.1
|
RNF111
|
ring finger protein 111
|
|
chr1_+_12464912
Show fit
|
0.02 |
ENST00000543766.2
|
VPS13D
|
vacuolar protein sorting 13 homolog D
|
|
chr12_+_71938837
Show fit
|
0.01 |
ENST00000333850.4
|
TPH2
|
tryptophan hydroxylase 2
|
|
chr13_-_27969295
Show fit
|
0.01 |
ENST00000381020.8
|
CDX2
|
caudal type homeobox 2
|
|
chr16_+_31527876
Show fit
|
0.01 |
ENST00000302312.9
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein
|
|
chr1_+_25272502
Show fit
|
0.01 |
ENST00000328664.9
|
RHD
|
Rh blood group D antigen
|
|
chr4_+_165873231
Show fit
|
0.01 |
ENST00000061240.7
|
TLL1
|
tolloid like 1
|
|
chr12_-_6124662
Show fit
|
0.01 |
ENST00000261405.10
|
VWF
|
von Willebrand factor
|
|
chr19_-_55038256
Show fit
|
0.01 |
ENST00000417454.5
ENST00000310373.7
ENST00000333884.2
|
GP6
|
glycoprotein VI platelet
|
|
chr11_-_26722051
Show fit
|
0.01 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12
|
|
chr9_-_97938157
Show fit
|
0.01 |
ENST00000616898.2
|
HEMGN
|
hemogen
|
|
chr1_+_154321076
Show fit
|
0.00 |
ENST00000324978.8
|
AQP10
|
aquaporin 10
|
|
chr11_+_75717811
Show fit
|
0.00 |
ENST00000198801.10
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2
|
|
chr12_-_21775581
Show fit
|
0.00 |
ENST00000537950.1
ENST00000665145.1
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8
|