Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
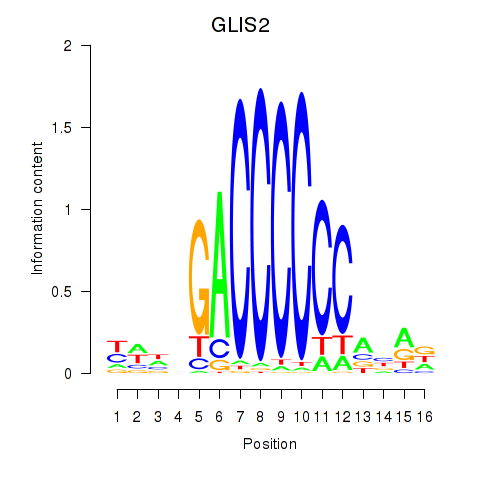
Results for GLIS2
Z-value: 0.99
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.9 | GLIS2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS2 | hg38_v1_chr16_+_4316052_4316075 | -0.56 | 1.2e-03 | Click! |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_35154914 | 5.79 |
ENST00000423817.7
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr19_+_35154715 | 5.68 |
ENST00000392218.6
ENST00000543307.5 ENST00000392219.7 ENST00000541435.6 ENST00000590686.5 ENST00000342879.7 ENST00000588699.5 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr6_-_2903300 | 3.48 |
ENST00000380698.5
|
SERPINB9
|
serpin family B member 9 |
| chr7_-_73719629 | 3.15 |
ENST00000395155.3
ENST00000395154.7 ENST00000395156.7 ENST00000222812.8 |
STX1A
|
syntaxin 1A |
| chr1_-_186680411 | 3.14 |
ENST00000367468.10
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 |
| chr6_-_39322688 | 2.98 |
ENST00000437525.3
|
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr2_+_233195433 | 2.54 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr1_-_28193873 | 2.25 |
ENST00000305392.3
ENST00000539896.1 |
PTAFR
|
platelet activating factor receptor |
| chr1_-_153375591 | 2.18 |
ENST00000368737.5
|
S100A12
|
S100 calcium binding protein A12 |
| chr2_+_37344594 | 2.13 |
ENST00000404976.5
ENST00000338415.8 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr1_-_6490564 | 2.12 |
ENST00000377725.5
ENST00000340850.10 |
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr19_+_44905785 | 2.04 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr19_+_48325323 | 1.98 |
ENST00000596315.5
|
EMP3
|
epithelial membrane protein 3 |
| chr22_+_22922594 | 1.96 |
ENST00000390331.3
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chrX_-_154374623 | 1.95 |
ENST00000369850.10
|
FLNA
|
filamin A |
| chr2_+_24049673 | 1.94 |
ENST00000380991.8
|
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr12_-_52848986 | 1.93 |
ENST00000304620.5
|
KRT78
|
keratin 78 |
| chr2_+_24049705 | 1.86 |
ENST00000380986.9
ENST00000452109.1 |
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr19_+_1407731 | 1.85 |
ENST00000592453.2
|
DAZAP1
|
DAZ associated protein 1 |
| chr19_+_11089446 | 1.74 |
ENST00000557933.5
ENST00000455727.6 ENST00000535915.5 ENST00000545707.5 ENST00000558518.6 ENST00000558013.5 |
LDLR
|
low density lipoprotein receptor |
| chr19_+_48321454 | 1.70 |
ENST00000599704.5
|
EMP3
|
epithelial membrane protein 3 |
| chr13_+_110307276 | 1.70 |
ENST00000360467.7
ENST00000650540.1 |
COL4A2
|
collagen type IV alpha 2 chain |
| chr19_+_53869384 | 1.67 |
ENST00000391769.2
|
MYADM
|
myeloid associated differentiation marker |
| chr19_+_44914833 | 1.67 |
ENST00000589078.1
ENST00000586638.5 |
APOC1
|
apolipoprotein C1 |
| chr14_+_94174284 | 1.67 |
ENST00000304338.8
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr5_-_177409535 | 1.64 |
ENST00000253496.4
|
F12
|
coagulation factor XII |
| chr9_+_113594118 | 1.62 |
ENST00000620489.1
|
RGS3
|
regulator of G protein signaling 3 |
| chrX_-_154371210 | 1.60 |
ENST00000369856.8
ENST00000422373.6 ENST00000360319.9 |
FLNA
|
filamin A |
| chr17_+_43398984 | 1.59 |
ENST00000320033.5
|
ARL4D
|
ADP ribosylation factor like GTPase 4D |
| chr8_+_27325516 | 1.56 |
ENST00000346049.10
ENST00000420218.3 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr13_-_20192928 | 1.53 |
ENST00000382848.5
|
GJB2
|
gap junction protein beta 2 |
| chr11_-_64878612 | 1.53 |
ENST00000320631.8
|
EHD1
|
EH domain containing 1 |
| chr19_+_44914588 | 1.50 |
ENST00000592535.6
|
APOC1
|
apolipoprotein C1 |
| chr5_-_132777215 | 1.46 |
ENST00000458488.2
|
SEPTIN8
|
septin 8 |
| chr19_+_44914247 | 1.45 |
ENST00000588750.5
ENST00000588802.5 |
APOC1
|
apolipoprotein C1 |
| chr1_-_155978427 | 1.42 |
ENST00000313667.8
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr3_-_48595267 | 1.42 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr21_+_33403391 | 1.42 |
ENST00000290219.11
ENST00000381995.5 |
IFNGR2
|
interferon gamma receptor 2 |
| chr19_+_44914702 | 1.42 |
ENST00000592885.5
ENST00000589781.1 |
APOC1
|
apolipoprotein C1 |
| chr1_-_231040218 | 1.34 |
ENST00000366654.5
|
FAM89A
|
family with sequence similarity 89 member A |
| chr19_+_17747698 | 1.30 |
ENST00000594202.5
ENST00000252771.11 |
FCHO1
|
FCH and mu domain containing endocytic adaptor 1 |
| chr16_-_90008988 | 1.30 |
ENST00000568662.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr11_+_844067 | 1.29 |
ENST00000397406.5
ENST00000409543.6 ENST00000525201.5 |
TSPAN4
|
tetraspanin 4 |
| chr22_+_22644475 | 1.29 |
ENST00000618722.4
ENST00000652219.1 ENST00000480559.6 ENST00000448514.2 ENST00000652249.1 ENST00000651213.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chr19_+_2249317 | 1.29 |
ENST00000221496.5
|
AMH
|
anti-Mullerian hormone |
| chr9_+_136952256 | 1.29 |
ENST00000371633.8
|
LCN12
|
lipocalin 12 |
| chr1_+_34782259 | 1.26 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chr16_+_30183595 | 1.25 |
ENST00000219150.10
ENST00000570045.5 ENST00000565497.5 ENST00000570244.5 |
CORO1A
|
coronin 1A |
| chr12_+_57460127 | 1.24 |
ENST00000532291.5
ENST00000543426.5 ENST00000546141.5 |
GLI1
|
GLI family zinc finger 1 |
| chr6_-_39322540 | 1.24 |
ENST00000425054.6
ENST00000373227.8 ENST00000373229.9 |
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr13_-_23433676 | 1.24 |
ENST00000682547.1
ENST00000455470.6 ENST00000382292.9 |
SACS
|
sacsin molecular chaperone |
| chr19_+_17747737 | 1.23 |
ENST00000600676.5
ENST00000600209.5 ENST00000596309.5 ENST00000598539.5 ENST00000597474.5 ENST00000593385.5 ENST00000598067.5 ENST00000593833.5 |
FCHO1
|
FCH and mu domain containing endocytic adaptor 1 |
| chr1_-_156705742 | 1.22 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr19_+_41755520 | 1.20 |
ENST00000199764.7
|
CEACAM6
|
CEA cell adhesion molecule 6 |
| chr21_-_30497160 | 1.17 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr20_+_46008900 | 1.16 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr19_-_42427379 | 1.14 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr5_+_169583636 | 1.14 |
ENST00000506574.5
ENST00000515224.5 ENST00000629457.2 ENST00000508247.5 ENST00000265295.9 ENST00000513941.5 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr20_-_23988779 | 1.12 |
ENST00000335694.4
|
GGTLC1
|
gamma-glutamyltransferase light chain 1 |
| chrX_-_30308333 | 1.12 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1 |
| chr3_-_48088824 | 1.12 |
ENST00000439356.2
ENST00000395734.7 ENST00000426837.6 |
MAP4
|
microtubule associated protein 4 |
| chr19_-_41353904 | 1.11 |
ENST00000221930.6
|
TGFB1
|
transforming growth factor beta 1 |
| chr1_-_156705764 | 1.09 |
ENST00000621784.4
ENST00000368220.1 |
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr6_-_39322968 | 1.08 |
ENST00000507712.5
|
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr9_+_136952896 | 1.08 |
ENST00000371632.7
|
LCN12
|
lipocalin 12 |
| chr1_-_151008365 | 1.07 |
ENST00000361936.9
ENST00000361738.11 |
MINDY1
|
MINDY lysine 48 deubiquitinase 1 |
| chr1_+_2019379 | 1.06 |
ENST00000638771.1
ENST00000640949.1 ENST00000640030.1 |
GABRD
|
gamma-aminobutyric acid type A receptor subunit delta |
| chr22_-_30289607 | 1.06 |
ENST00000404953.7
|
CASTOR1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr20_+_37346128 | 1.04 |
ENST00000373578.7
|
SRC
|
SRC proto-oncogene, non-receptor tyrosine kinase |
| chr16_+_57245229 | 1.04 |
ENST00000219204.8
|
ARL2BP
|
ADP ribosylation factor like GTPase 2 binding protein |
| chr11_+_111245725 | 1.04 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr2_+_219627622 | 1.01 |
ENST00000358055.8
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr17_+_6444441 | 1.01 |
ENST00000250056.12
ENST00000571373.5 ENST00000570337.6 ENST00000572595.6 ENST00000572447.6 ENST00000576056.5 |
PIMREG
|
PICALM interacting mitotic regulator |
| chr22_-_27801712 | 1.00 |
ENST00000302326.5
|
MN1
|
MN1 proto-oncogene, transcriptional regulator |
| chr3_+_111859284 | 1.00 |
ENST00000498699.5
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr2_+_219627650 | 1.00 |
ENST00000317151.7
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr21_+_33403466 | 0.99 |
ENST00000405436.5
|
IFNGR2
|
interferon gamma receptor 2 |
| chr1_-_156705575 | 0.98 |
ENST00000368222.8
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr5_-_1523900 | 0.96 |
ENST00000283415.4
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1 |
| chr16_-_28506826 | 0.96 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr16_+_70646536 | 0.94 |
ENST00000288098.6
|
IL34
|
interleukin 34 |
| chr11_-_1622093 | 0.94 |
ENST00000616115.1
ENST00000399682.1 |
KRTAP5-4
|
keratin associated protein 5-4 |
| chr10_-_133336862 | 0.92 |
ENST00000368555.3
ENST00000252939.9 ENST00000368558.1 |
CALY
|
calcyon neuron specific vesicular protein |
| chr3_+_111859180 | 0.92 |
ENST00000412622.5
ENST00000431670.7 |
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr19_-_37906646 | 0.89 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chr11_+_450255 | 0.89 |
ENST00000308020.6
|
PTDSS2
|
phosphatidylserine synthase 2 |
| chr19_+_3506355 | 0.85 |
ENST00000652521.1
|
FZR1
|
fizzy and cell division cycle 20 related 1 |
| chr11_-_627173 | 0.85 |
ENST00000176195.4
|
SCT
|
secretin |
| chr9_+_100427123 | 0.83 |
ENST00000395067.7
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr20_+_64084995 | 0.83 |
ENST00000349451.3
|
OPRL1
|
opioid related nociceptin receptor 1 |
| chr13_+_42272134 | 0.81 |
ENST00000025301.4
|
AKAP11
|
A-kinase anchoring protein 11 |
| chr2_-_89010515 | 0.81 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr19_-_37906588 | 0.80 |
ENST00000447313.7
|
WDR87
|
WD repeat domain 87 |
| chr9_+_100427254 | 0.80 |
ENST00000374885.5
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr1_+_35079374 | 0.80 |
ENST00000359858.9
ENST00000373330.1 |
ZMYM1
|
zinc finger MYM-type containing 1 |
| chr9_+_123356189 | 0.80 |
ENST00000373631.8
|
CRB2
|
crumbs cell polarity complex component 2 |
| chr3_+_111859796 | 0.79 |
ENST00000393925.7
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr22_-_39244969 | 0.78 |
ENST00000331163.11
|
PDGFB
|
platelet derived growth factor subunit B |
| chr6_-_150069066 | 0.77 |
ENST00000438272.2
ENST00000367339.7 |
ULBP3
|
UL16 binding protein 3 |
| chr19_-_45902594 | 0.77 |
ENST00000322217.6
|
MYPOP
|
Myb related transcription factor, partner of profilin |
| chr10_+_104254915 | 0.77 |
ENST00000445155.5
|
GSTO1
|
glutathione S-transferase omega 1 |
| chr22_+_29073112 | 0.75 |
ENST00000327813.9
ENST00000407188.5 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr16_+_57245336 | 0.75 |
ENST00000562023.5
ENST00000563234.1 |
ARL2BP
|
ADP ribosylation factor like GTPase 2 binding protein |
| chr1_-_155978144 | 0.74 |
ENST00000313695.11
ENST00000497907.5 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr10_+_104254867 | 0.73 |
ENST00000369713.10
|
GSTO1
|
glutathione S-transferase omega 1 |
| chr5_-_141680822 | 0.69 |
ENST00000504448.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr20_+_54208072 | 0.69 |
ENST00000371419.7
|
PFDN4
|
prefoldin subunit 4 |
| chr17_-_42979993 | 0.69 |
ENST00000409103.5
ENST00000360221.8 ENST00000454303.1 ENST00000591916.7 ENST00000451885.3 |
PTGES3L-AARSD1
PTGES3L
|
PTGES3L-AARSD1 readthrough prostaglandin E synthase 3 like |
| chr16_+_27066919 | 0.67 |
ENST00000505035.3
|
C16orf82
|
chromosome 16 open reading frame 82 |
| chr12_+_7155867 | 0.67 |
ENST00000535313.2
ENST00000331148.5 |
CLSTN3
|
calsyntenin 3 |
| chr11_-_72642407 | 0.67 |
ENST00000376450.7
|
PDE2A
|
phosphodiesterase 2A |
| chr19_-_36152427 | 0.66 |
ENST00000589154.1
ENST00000292907.8 |
COX7A1
|
cytochrome c oxidase subunit 7A1 |
| chr20_-_31845593 | 0.65 |
ENST00000375978.5
|
FOXS1
|
forkhead box S1 |
| chr11_-_72642450 | 0.65 |
ENST00000444035.6
ENST00000544570.5 |
PDE2A
|
phosphodiesterase 2A |
| chr19_-_4517600 | 0.65 |
ENST00000301286.4
|
PLIN4
|
perilipin 4 |
| chr2_-_135876382 | 0.65 |
ENST00000264156.3
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr19_-_40750302 | 0.65 |
ENST00000598485.6
ENST00000378313.7 ENST00000470681.5 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr16_+_69424634 | 0.64 |
ENST00000515314.6
ENST00000561792.6 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B |
| chr19_+_1407653 | 0.63 |
ENST00000587079.5
|
DAZAP1
|
DAZ associated protein 1 |
| chr1_+_150982268 | 0.62 |
ENST00000368947.9
|
ANXA9
|
annexin A9 |
| chr12_-_110689105 | 0.62 |
ENST00000242607.13
|
HVCN1
|
hydrogen voltage gated channel 1 |
| chr9_-_35112379 | 0.62 |
ENST00000488109.6
|
FAM214B
|
family with sequence similarity 214 member B |
| chr14_-_106211453 | 0.62 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr1_-_201115372 | 0.62 |
ENST00000458416.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr19_+_3185911 | 0.61 |
ENST00000246117.9
ENST00000588428.5 |
NCLN
|
nicalin |
| chr8_-_86069662 | 0.61 |
ENST00000276616.3
|
PSKH2
|
protein serine kinase H2 |
| chr21_+_46001300 | 0.61 |
ENST00000612273.2
ENST00000682634.1 |
COL6A1
|
collagen type VI alpha 1 chain |
| chr19_+_55339867 | 0.61 |
ENST00000255613.8
|
KMT5C
|
lysine methyltransferase 5C |
| chr17_+_42017020 | 0.61 |
ENST00000307641.9
|
NKIRAS2
|
NFKB inhibitor interacting Ras like 2 |
| chr3_+_38165484 | 0.60 |
ENST00000446845.5
ENST00000311806.8 |
OXSR1
|
oxidative stress responsive kinase 1 |
| chr3_-_43621897 | 0.60 |
ENST00000444344.5
ENST00000456438.5 ENST00000350459.8 ENST00000396091.7 ENST00000451430.6 ENST00000428472.5 ENST00000292246.8 ENST00000414522.6 |
ANO10
|
anoctamin 10 |
| chr8_+_38386433 | 0.60 |
ENST00000297720.9
ENST00000524874.5 ENST00000379957.9 ENST00000523983.6 |
LETM2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr11_+_68903849 | 0.59 |
ENST00000675615.1
ENST00000255078.8 |
IGHMBP2
|
immunoglobulin mu DNA binding protein 2 |
| chr5_-_132777404 | 0.58 |
ENST00000296873.11
|
SEPTIN8
|
septin 8 |
| chr12_-_57547224 | 0.58 |
ENST00000678322.1
|
DCTN2
|
dynactin subunit 2 |
| chr17_+_7307579 | 0.57 |
ENST00000572815.5
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr19_+_45079195 | 0.57 |
ENST00000591607.1
ENST00000591747.5 ENST00000270257.9 ENST00000391951.2 ENST00000587566.5 |
GEMIN7
MARK4
|
gem nuclear organelle associated protein 7 microtubule affinity regulating kinase 4 |
| chr7_+_101154981 | 0.57 |
ENST00000646560.1
|
AP1S1
|
adaptor related protein complex 1 subunit sigma 1 |
| chr1_+_2019324 | 0.56 |
ENST00000638411.1
ENST00000378585.7 ENST00000640067.1 |
GABRD
|
gamma-aminobutyric acid type A receptor subunit delta |
| chr1_+_65147514 | 0.56 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr20_+_56533246 | 0.55 |
ENST00000371325.1
|
FAM209B
|
family with sequence similarity 209 member B |
| chr12_-_57547121 | 0.55 |
ENST00000550954.5
ENST00000434715.7 ENST00000678505.1 ENST00000543672.6 ENST00000546670.5 ENST00000548249.6 |
DCTN2
|
dynactin subunit 2 |
| chr17_+_18108706 | 0.55 |
ENST00000615845.4
|
MYO15A
|
myosin XVA |
| chr19_-_40285277 | 0.55 |
ENST00000579047.5
ENST00000392038.7 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr7_-_727242 | 0.55 |
ENST00000537384.6
ENST00000417852.5 |
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr14_+_99684283 | 0.54 |
ENST00000261835.8
|
CYP46A1
|
cytochrome P450 family 46 subfamily A member 1 |
| chr22_+_50674879 | 0.54 |
ENST00000262795.6
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr11_-_2423012 | 0.54 |
ENST00000533881.5
ENST00000533060.5 ENST00000528453.1 ENST00000155858.10 |
TRPM5
|
transient receptor potential cation channel subfamily M member 5 |
| chr17_+_18108747 | 0.53 |
ENST00000647165.2
|
MYO15A
|
myosin XVA |
| chr19_+_1275508 | 0.52 |
ENST00000409293.6
|
FAM174C
|
family with sequence similarity 174 member C |
| chr7_-_727611 | 0.52 |
ENST00000403562.5
|
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr5_+_150671588 | 0.52 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr1_+_160081529 | 0.51 |
ENST00000368088.4
|
KCNJ9
|
potassium inwardly rectifying channel subfamily J member 9 |
| chr9_+_124869126 | 0.51 |
ENST00000259477.6
|
ARPC5L
|
actin related protein 2/3 complex subunit 5 like |
| chr13_+_46553157 | 0.51 |
ENST00000311191.10
ENST00000389797.8 ENST00000389798.7 |
LRCH1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr20_-_31723491 | 0.50 |
ENST00000676582.1
ENST00000422920.2 |
BCL2L1
|
BCL2 like 1 |
| chr7_-_712940 | 0.50 |
ENST00000544935.5
ENST00000430040.5 ENST00000456696.2 ENST00000406797.5 |
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr14_-_105601728 | 0.49 |
ENST00000641420.1
ENST00000390541.2 |
IGHE
|
immunoglobulin heavy constant epsilon |
| chr19_+_39413528 | 0.49 |
ENST00000438123.5
ENST00000409797.6 ENST00000451354.6 |
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr19_+_53909263 | 0.49 |
ENST00000391767.6
|
CACNG7
|
calcium voltage-gated channel auxiliary subunit gamma 7 |
| chr17_-_5234801 | 0.48 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr5_-_132777866 | 0.48 |
ENST00000448933.5
|
SEPTIN8
|
septin 8 |
| chr10_+_96304392 | 0.48 |
ENST00000630152.1
|
DNTT
|
DNA nucleotidylexotransferase |
| chr5_-_132777371 | 0.48 |
ENST00000620483.4
|
SEPTIN8
|
septin 8 |
| chr10_+_96304425 | 0.47 |
ENST00000371174.5
|
DNTT
|
DNA nucleotidylexotransferase |
| chr8_+_1823918 | 0.47 |
ENST00000349830.8
|
ARHGEF10
|
Rho guanine nucleotide exchange factor 10 |
| chr16_+_3020359 | 0.47 |
ENST00000341627.5
ENST00000326577.9 ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
TNF receptor superfamily member 12A |
| chr19_+_14529580 | 0.46 |
ENST00000215567.10
|
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr7_-_712437 | 0.46 |
ENST00000360274.8
|
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr3_-_28348924 | 0.45 |
ENST00000414162.5
ENST00000420543.6 |
AZI2
|
5-azacytidine induced 2 |
| chr1_-_23800402 | 0.45 |
ENST00000374497.7
ENST00000425913.5 |
GALE
|
UDP-galactose-4-epimerase |
| chr19_+_3506251 | 0.45 |
ENST00000441788.7
|
FZR1
|
fizzy and cell division cycle 20 related 1 |
| chr3_-_177197210 | 0.45 |
ENST00000431421.5
ENST00000422066.5 ENST00000413084.5 ENST00000673974.1 ENST00000422442.6 |
TBL1XR1
|
TBL1X receptor 1 |
| chr9_-_35958154 | 0.45 |
ENST00000341959.2
|
OR2S2
|
olfactory receptor family 2 subfamily S member 2 |
| chr12_-_54584302 | 0.45 |
ENST00000553113.5
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A |
| chr12_-_48957365 | 0.44 |
ENST00000398092.4
ENST00000539611.1 |
ENSG00000272822.2
ARF3
|
novel protein ADP ribosylation factor 3 |
| chr19_-_3772211 | 0.44 |
ENST00000555978.5
ENST00000555633.3 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr15_-_34343112 | 0.44 |
ENST00000557912.1
ENST00000328848.6 |
NOP10
|
NOP10 ribonucleoprotein |
| chr19_+_13764502 | 0.43 |
ENST00000040663.8
ENST00000319545.12 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr14_-_56810380 | 0.43 |
ENST00000672125.1
ENST00000673481.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr19_-_40285395 | 0.43 |
ENST00000424901.5
ENST00000578123.5 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr19_-_38812936 | 0.42 |
ENST00000307751.9
ENST00000594209.1 |
LGALS4
|
galectin 4 |
| chr6_-_70303070 | 0.42 |
ENST00000370496.3
ENST00000357250.11 |
COL9A1
|
collagen type IX alpha 1 chain |
| chr15_+_90935277 | 0.41 |
ENST00000418476.2
|
UNC45A
|
unc-45 myosin chaperone A |
| chr17_-_41836193 | 0.41 |
ENST00000435506.7
ENST00000415460.5 |
NT5C3B
|
5'-nucleotidase, cytosolic IIIB |
| chr2_-_196926717 | 0.41 |
ENST00000409475.5
ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr19_+_10871516 | 0.41 |
ENST00000327064.9
ENST00000588947.5 |
CARM1
|
coactivator associated arginine methyltransferase 1 |
| chr19_+_44751251 | 0.40 |
ENST00000444487.1
|
BCL3
|
BCL3 transcription coactivator |
| chr19_+_55605639 | 0.40 |
ENST00000568956.2
|
ZNF865
|
zinc finger protein 865 |
| chr2_-_15561305 | 0.40 |
ENST00000281513.10
|
NBAS
|
NBAS subunit of NRZ tethering complex |
| chr17_+_82458174 | 0.40 |
ENST00000579198.5
ENST00000390006.8 ENST00000580296.5 |
NARF
|
nuclear prelamin A recognition factor |
| chr1_-_201154418 | 0.40 |
ENST00000435310.5
ENST00000485839.6 |
TMEM9
|
transmembrane protein 9 |
| chr1_-_114757971 | 0.39 |
ENST00000261443.9
ENST00000534699.5 ENST00000339438.10 ENST00000358528.9 ENST00000529046.5 ENST00000525970.5 ENST00000369530.5 ENST00000530886.5 ENST00000610726.4 |
CSDE1
|
cold shock domain containing E1 |
| chr14_-_77498808 | 0.38 |
ENST00000342219.9
ENST00000493585.5 ENST00000554801.2 |
ISM2
|
isthmin 2 |
| chr5_-_170389634 | 0.38 |
ENST00000521859.1
|
KCNMB1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr2_+_9843426 | 0.38 |
ENST00000625239.2
ENST00000263663.10 |
TAF1B
|
TATA-box binding protein associated factor, RNA polymerase I subunit B |
| chr3_-_11846772 | 0.38 |
ENST00000455809.5
ENST00000444133.6 ENST00000273037.9 ENST00000630288.1 |
TAMM41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr15_-_79090760 | 0.38 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr12_-_109833373 | 0.38 |
ENST00000261740.7
|
TRPV4
|
transient receptor potential cation channel subfamily V member 4 |
| chrX_+_46573757 | 0.38 |
ENST00000276055.4
|
CHST7
|
carbohydrate sulfotransferase 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.5 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 1.5 | 6.0 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 1.2 | 3.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.2 | 3.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 1.1 | 3.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.7 | 2.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.6 | 2.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.6 | 3.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.6 | 1.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 2.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.5 | 1.6 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.5 | 1.5 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 0.5 | 2.3 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.4 | 1.7 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.4 | 1.7 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.4 | 2.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 1.3 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.4 | 1.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.4 | 1.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 3.8 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.4 | 1.1 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.3 | 1.0 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.3 | 1.5 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.3 | 0.8 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.3 | 1.1 | GO:0052509 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.3 | 0.8 | GO:0014028 | notochord formation(GO:0014028) |
| 0.3 | 1.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 | 3.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 2.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.0 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) cellular response to progesterone stimulus(GO:0071393) |
| 0.2 | 0.6 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 0.2 | 1.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 0.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 3.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.5 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.6 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 1.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 3.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) negative regulation of actin nucleation(GO:0051126) |
| 0.1 | 0.4 | GO:0034970 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.4 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.4 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 0.4 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 1.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 3.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 2.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.5 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.4 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.0 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 2.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.2 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 1.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.1 | GO:0060516 | primary prostatic bud elongation(GO:0060516) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 1.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 1.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.9 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 1.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 2.2 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.4 | GO:0060762 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 3.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.3 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.6 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.3 | GO:0051754 | female meiosis chromosome segregation(GO:0016321) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 1.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) |
| 0.0 | 1.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 1.9 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.4 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 2.2 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.7 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 1.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 1.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.3 | GO:0071787 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 4.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.4 | GO:0021924 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 2.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0070315 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.7 | 3.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.6 | 1.7 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.5 | 2.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 1.5 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 1.4 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.2 | 5.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 3.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 2.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 2.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.4 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 3.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 3.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 3.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.9 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.7 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.7 | 8.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.7 | 3.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 3.1 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.6 | 2.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.6 | 2.3 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.4 | 2.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.4 | 1.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 1.5 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 3.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.8 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.2 | 1.0 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 1.1 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 3.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 2.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 3.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 1.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 1.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 0.5 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 1.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.4 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 10.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.9 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 2.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.4 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.5 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 1.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 2.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 2.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.9 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.1 | 0.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 3.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 3.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 1.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 2.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 1.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 3.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0030942 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.6 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 4.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 4.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 4.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 3.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 3.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 2.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |