Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for GLIS3
Z-value: 0.35
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.24 | GLIS3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS3 | hg38_v1_chr9_-_4300049_4300120, hg38_v1_chr9_-_4299547_4299597 | 0.49 | 5.8e-03 | Click! |
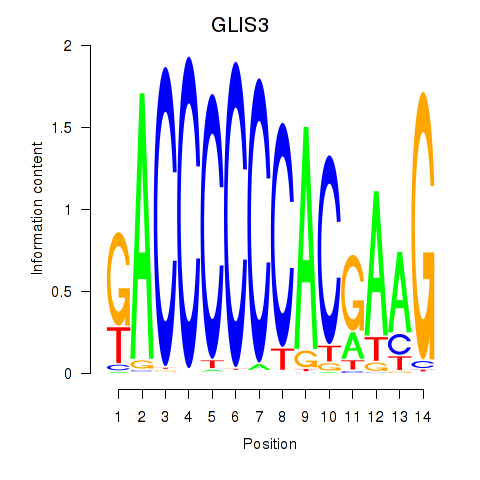
Activity profile of GLIS3 motif
Sorted Z-values of GLIS3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_107044689 | 1.07 |
ENST00000265717.5
|
PRKAR2B
|
protein kinase cAMP-dependent type II regulatory subunit beta |
| chr5_+_172959511 | 0.65 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26 like 1 |
| chr6_+_24494939 | 0.51 |
ENST00000348925.2
ENST00000357578.8 |
ALDH5A1
|
aldehyde dehydrogenase 5 family member A1 |
| chr6_+_89081787 | 0.43 |
ENST00000354922.3
|
PNRC1
|
proline rich nuclear receptor coactivator 1 |
| chr15_+_83447411 | 0.36 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr1_-_157138474 | 0.32 |
ENST00000326786.4
|
ETV3
|
ETS variant transcription factor 3 |
| chr1_-_19799872 | 0.32 |
ENST00000294543.11
|
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr17_-_79952007 | 0.26 |
ENST00000574241.6
|
TBC1D16
|
TBC1 domain family member 16 |
| chr17_-_5111836 | 0.25 |
ENST00000575898.5
|
ZNF232
|
zinc finger protein 232 |
| chr6_-_44265541 | 0.24 |
ENST00000619360.6
|
NFKBIE
|
NFKB inhibitor epsilon |
| chr8_-_29263063 | 0.23 |
ENST00000524189.6
|
KIF13B
|
kinesin family member 13B |
| chr16_+_84145256 | 0.21 |
ENST00000378553.10
|
DNAAF1
|
dynein axonemal assembly factor 1 |
| chr21_-_44910630 | 0.21 |
ENST00000320216.10
ENST00000397852.5 |
ITGB2
|
integrin subunit beta 2 |
| chr20_+_21126037 | 0.21 |
ENST00000611685.4
ENST00000616848.4 |
KIZ
|
kizuna centrosomal protein |
| chr8_+_11769639 | 0.21 |
ENST00000436750.7
|
NEIL2
|
nei like DNA glycosylase 2 |
| chr20_+_21125999 | 0.21 |
ENST00000620891.4
|
KIZ
|
kizuna centrosomal protein |
| chr20_+_21125981 | 0.20 |
ENST00000619574.4
|
KIZ
|
kizuna centrosomal protein |
| chr22_+_22644475 | 0.20 |
ENST00000618722.4
ENST00000652219.1 ENST00000480559.6 ENST00000448514.2 ENST00000652249.1 ENST00000651213.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chr18_-_48950960 | 0.20 |
ENST00000262158.8
|
SMAD7
|
SMAD family member 7 |
| chr20_+_21126074 | 0.20 |
ENST00000619189.5
|
KIZ
|
kizuna centrosomal protein |
| chr11_+_818906 | 0.18 |
ENST00000336615.9
|
PNPLA2
|
patatin like phospholipase domain containing 2 |
| chr8_+_11769696 | 0.18 |
ENST00000455213.6
ENST00000403422.7 ENST00000528323.5 ENST00000284503.7 |
NEIL2
|
nei like DNA glycosylase 2 |
| chr3_+_173398438 | 0.16 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1 |
| chr16_-_3235143 | 0.15 |
ENST00000414144.7
ENST00000431561.7 ENST00000396870.8 |
ZNF200
|
zinc finger protein 200 |
| chr1_-_53945584 | 0.15 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr12_+_1820255 | 0.15 |
ENST00000543818.5
|
LRTM2
|
leucine rich repeats and transmembrane domains 2 |
| chr19_-_49325181 | 0.15 |
ENST00000454748.7
ENST00000335875.9 ENST00000598828.1 |
SLC6A16
|
solute carrier family 6 member 16 |
| chr1_-_53945567 | 0.14 |
ENST00000371378.6
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr1_+_107056656 | 0.13 |
ENST00000370078.2
|
PRMT6
|
protein arginine methyltransferase 6 |
| chr19_+_43984167 | 0.13 |
ENST00000611002.4
ENST00000270014.7 ENST00000591532.5 ENST00000407951.6 ENST00000590615.5 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chr6_+_44223553 | 0.12 |
ENST00000371740.10
ENST00000371755.9 ENST00000643869.1 ENST00000371731.6 |
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr15_-_43590155 | 0.12 |
ENST00000453080.5
ENST00000360135.8 ENST00000360301.8 ENST00000417085.2 ENST00000431962.1 ENST00000334933.8 ENST00000381879.8 ENST00000420765.5 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr17_+_50835578 | 0.12 |
ENST00000311378.5
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr8_-_73878816 | 0.11 |
ENST00000602593.6
ENST00000651945.1 ENST00000419880.7 ENST00000517608.5 ENST00000650817.1 |
UBE2W
|
ubiquitin conjugating enzyme E2 W |
| chr9_-_113299196 | 0.11 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr19_-_55580829 | 0.10 |
ENST00000592239.1
ENST00000325421.7 |
ZNF579
|
zinc finger protein 579 |
| chr17_+_7888783 | 0.10 |
ENST00000330494.12
ENST00000358181.8 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr17_+_76000906 | 0.10 |
ENST00000448471.2
|
CDK3
|
cyclin dependent kinase 3 |
| chr19_+_40775154 | 0.10 |
ENST00000594436.5
ENST00000597784.5 |
MIA
|
MIA SH3 domain containing |
| chr2_+_219514477 | 0.10 |
ENST00000347842.8
ENST00000358078.5 |
ASIC4
|
acid sensing ion channel subunit family member 4 |
| chr6_+_44223770 | 0.10 |
ENST00000652453.1
ENST00000393841.6 ENST00000371724.6 ENST00000642777.1 ENST00000645692.1 |
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr11_-_68903796 | 0.10 |
ENST00000362034.7
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr1_+_2019324 | 0.10 |
ENST00000638411.1
ENST00000378585.7 ENST00000640067.1 |
GABRD
|
gamma-aminobutyric acid type A receptor subunit delta |
| chr19_-_37906646 | 0.09 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chr4_-_83334782 | 0.09 |
ENST00000681769.1
ENST00000513463.1 ENST00000311412.10 |
HPSE
|
heparanase |
| chr16_+_89686661 | 0.09 |
ENST00000505473.5
ENST00000353379.12 ENST00000625631.1 ENST00000564192.5 |
CDK10
|
cyclin dependent kinase 10 |
| chr19_-_37906588 | 0.09 |
ENST00000447313.7
|
WDR87
|
WD repeat domain 87 |
| chr1_+_2019379 | 0.09 |
ENST00000638771.1
ENST00000640949.1 ENST00000640030.1 |
GABRD
|
gamma-aminobutyric acid type A receptor subunit delta |
| chr19_+_13764502 | 0.09 |
ENST00000040663.8
ENST00000319545.12 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr17_+_50834581 | 0.09 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr17_-_35119733 | 0.09 |
ENST00000460118.6
ENST00000335858.11 |
RAD51D
|
RAD51 paralog D |
| chr17_-_35119801 | 0.09 |
ENST00000592577.5
ENST00000590016.5 ENST00000345365.11 |
RAD51D
|
RAD51 paralog D |
| chr22_-_50526337 | 0.08 |
ENST00000651490.1
ENST00000543927.6 |
TYMP
SCO2
|
thymidine phosphorylase synthesis of cytochrome C oxidase 2 |
| chr12_+_109139397 | 0.08 |
ENST00000377854.9
ENST00000377848.7 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr10_-_74163 | 0.07 |
ENST00000564130.2
|
TUBB8
|
tubulin beta 8 class VIII |
| chr20_-_46089905 | 0.07 |
ENST00000372291.3
ENST00000290231.11 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr19_-_10502745 | 0.06 |
ENST00000393623.6
|
KEAP1
|
kelch like ECH associated protein 1 |
| chr5_-_132777229 | 0.06 |
ENST00000378721.8
ENST00000378719.7 ENST00000378701.5 |
SEPTIN8
|
septin 8 |
| chr19_-_3061403 | 0.06 |
ENST00000586839.1
|
TLE5
|
TLE family member 5, transcriptional modulator |
| chr17_-_75182536 | 0.06 |
ENST00000578238.2
|
SUMO2
|
small ubiquitin like modifier 2 |
| chr19_-_42255119 | 0.06 |
ENST00000222329.9
ENST00000594664.1 |
ERF
ENSG00000268643.2
|
ETS2 repressor factor novel protein |
| chr8_+_117134989 | 0.05 |
ENST00000456015.7
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr21_+_36699100 | 0.05 |
ENST00000290399.11
|
SIM2
|
SIM bHLH transcription factor 2 |
| chr8_+_117135259 | 0.05 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chrX_+_119236245 | 0.05 |
ENST00000535419.2
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr5_+_172959416 | 0.05 |
ENST00000265100.6
ENST00000519239.5 |
RPL26L1
|
ribosomal protein L26 like 1 |
| chr19_-_10503186 | 0.05 |
ENST00000592055.2
ENST00000171111.10 |
KEAP1
|
kelch like ECH associated protein 1 |
| chr17_+_4771878 | 0.05 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr17_+_5486285 | 0.04 |
ENST00000576988.1
ENST00000576570.5 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr9_-_136764515 | 0.04 |
ENST00000316144.6
|
LCN15
|
lipocalin 15 |
| chr3_-_25783434 | 0.04 |
ENST00000396649.7
ENST00000280700.10 ENST00000428257.5 |
NGLY1
|
N-glycanase 1 |
| chr1_-_246931892 | 0.04 |
ENST00000648844.2
|
AHCTF1
|
AT-hook containing transcription factor 1 |
| chr19_+_1275508 | 0.03 |
ENST00000409293.6
|
FAM174C
|
family with sequence similarity 174 member C |
| chr1_-_160285120 | 0.03 |
ENST00000368072.10
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chrX_+_119236274 | 0.03 |
ENST00000217971.8
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr18_+_3451647 | 0.03 |
ENST00000345133.9
ENST00000330513.10 ENST00000549546.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr10_+_61901678 | 0.03 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr3_-_25783381 | 0.03 |
ENST00000308710.9
ENST00000676225.1 |
NGLY1
|
N-glycanase 1 |
| chr17_-_75182949 | 0.03 |
ENST00000314523.7
ENST00000420826.7 |
SUMO2
|
small ubiquitin like modifier 2 |
| chr9_-_95516959 | 0.03 |
ENST00000437951.6
ENST00000430669.6 ENST00000468211.6 |
PTCH1
|
patched 1 |
| chr10_-_125161019 | 0.03 |
ENST00000411419.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr12_+_54008961 | 0.02 |
ENST00000040584.6
|
HOXC8
|
homeobox C8 |
| chr18_+_3451585 | 0.02 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr1_-_42335189 | 0.02 |
ENST00000361776.5
ENST00000445886.5 ENST00000361346.6 |
FOXJ3
|
forkhead box J3 |
| chr19_+_11798514 | 0.02 |
ENST00000323169.10
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr1_-_52552994 | 0.02 |
ENST00000355809.4
ENST00000528642.5 ENST00000470626.1 ENST00000257177.9 ENST00000371544.7 |
TUT4
|
terminal uridylyl transferase 4 |
| chr1_-_42335869 | 0.01 |
ENST00000372573.5
|
FOXJ3
|
forkhead box J3 |
| chr6_+_33204645 | 0.01 |
ENST00000374662.4
|
HSD17B8
|
hydroxysteroid 17-beta dehydrogenase 8 |
| chr3_-_142963663 | 0.01 |
ENST00000340634.6
|
PAQR9
|
progestin and adipoQ receptor family member 9 |
| chr16_+_69187125 | 0.01 |
ENST00000336278.8
|
SNTB2
|
syntrophin beta 2 |
| chr4_+_163343882 | 0.00 |
ENST00000338566.8
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr19_+_12945839 | 0.00 |
ENST00000586534.6
ENST00000316856.7 ENST00000592268.5 |
RAD23A
|
RAD23 homolog A, nucleotide excision repair protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.5 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:2000320 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.4 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0036155 | positive regulation of triglyceride catabolic process(GO:0010898) acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.0 | 0.1 | GO:0016154 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |