Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
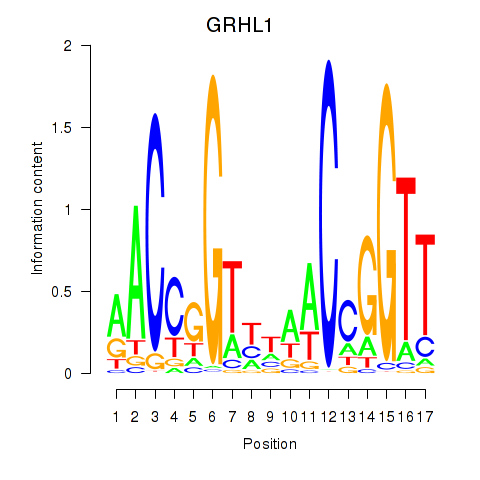
Results for GRHL1
Z-value: 0.69
Transcription factors associated with GRHL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GRHL1
|
ENSG00000134317.18 | GRHL1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GRHL1 | hg38_v1_chr2_+_9951653_9951734 | 0.63 | 1.8e-04 | Click! |
Activity profile of GRHL1 motif
Sorted Z-values of GRHL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GRHL1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_102642791 | 1.75 |
ENST00000340457.8
|
UPK3BL1
|
uroplakin 3B like 1 |
| chr7_-_102543849 | 1.67 |
ENST00000644544.1
|
UPK3BL2
|
uroplakin 3B like 2 |
| chr11_-_67674725 | 1.40 |
ENST00000525827.6
ENST00000673966.1 ENST00000673873.1 |
ALDH3B2
|
aldehyde dehydrogenase 3 family member B2 |
| chr11_-_67674606 | 1.33 |
ENST00000674110.1
ENST00000349015.7 |
ALDH3B2
|
aldehyde dehydrogenase 3 family member B2 |
| chr11_+_706595 | 1.15 |
ENST00000531348.5
ENST00000530636.5 |
EPS8L2
|
EPS8 like 2 |
| chr1_-_24143112 | 1.15 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr7_+_76510608 | 1.10 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr7_+_76510528 | 1.08 |
ENST00000334348.8
|
UPK3B
|
uroplakin 3B |
| chr2_-_85414039 | 1.08 |
ENST00000447219.6
ENST00000409670.5 ENST00000409724.5 |
CAPG
|
capping actin protein, gelsolin like |
| chr7_+_76510516 | 1.07 |
ENST00000257632.9
|
UPK3B
|
uroplakin 3B |
| chr1_-_201399302 | 0.98 |
ENST00000633953.1
ENST00000391967.7 |
LAD1
|
ladinin 1 |
| chr7_-_80922354 | 0.96 |
ENST00000419255.6
|
SEMA3C
|
semaphorin 3C |
| chr14_-_80211268 | 0.93 |
ENST00000556811.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr3_+_101849505 | 0.86 |
ENST00000326151.9
ENST00000326172.9 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr10_+_103277129 | 0.83 |
ENST00000369849.9
|
INA
|
internexin neuronal intermediate filament protein alpha |
| chr2_+_95274439 | 0.83 |
ENST00000317620.14
ENST00000403131.6 ENST00000317668.8 |
PROM2
|
prominin 2 |
| chr14_-_80211472 | 0.80 |
ENST00000557125.1
ENST00000438257.9 ENST00000422005.7 |
DIO2
|
iodothyronine deiodinase 2 |
| chr11_+_65787056 | 0.76 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr2_+_113058637 | 0.76 |
ENST00000346807.7
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr9_+_5450503 | 0.74 |
ENST00000381573.8
ENST00000381577.4 |
CD274
|
CD274 molecule |
| chr8_-_129786617 | 0.73 |
ENST00000276708.9
|
GSDMC
|
gasdermin C |
| chr20_-_62427528 | 0.70 |
ENST00000252998.2
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr11_+_76782250 | 0.57 |
ENST00000533752.1
ENST00000612930.1 |
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr5_+_181040260 | 0.55 |
ENST00000515271.1
ENST00000327705.14 |
BTNL9
|
butyrophilin like 9 |
| chr1_-_28058087 | 0.54 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr11_+_119168188 | 0.53 |
ENST00000454811.5
ENST00000409265.8 ENST00000449394.5 |
NLRX1
|
NLR family member X1 |
| chr11_+_119168705 | 0.53 |
ENST00000409109.6
ENST00000409991.5 ENST00000292199.6 |
NLRX1
|
NLR family member X1 |
| chr1_-_153608136 | 0.52 |
ENST00000368703.6
|
S100A16
|
S100 calcium binding protein A16 |
| chr1_+_34782259 | 0.51 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chr15_+_41838839 | 0.50 |
ENST00000458483.4
|
PLA2G4B
|
phospholipase A2 group IVB |
| chr19_-_35490456 | 0.49 |
ENST00000338897.4
ENST00000484218.6 |
KRTDAP
|
keratinocyte differentiation associated protein |
| chr20_-_25339731 | 0.48 |
ENST00000450393.5
ENST00000491682.5 |
ABHD12
|
abhydrolase domain containing 12, lysophospholipase |
| chr9_+_69121259 | 0.47 |
ENST00000643713.1
ENST00000606364.5 |
TJP2
|
tight junction protein 2 |
| chr18_-_37565825 | 0.47 |
ENST00000603232.6
|
CELF4
|
CUGBP Elav-like family member 4 |
| chr19_-_15479469 | 0.46 |
ENST00000292609.8
ENST00000340880.5 |
PGLYRP2
|
peptidoglycan recognition protein 2 |
| chr11_+_706117 | 0.45 |
ENST00000533256.5
ENST00000614442.4 |
EPS8L2
|
EPS8 like 2 |
| chr9_+_69123009 | 0.42 |
ENST00000647986.1
|
TJP2
|
tight junction protein 2 |
| chr15_+_41559189 | 0.42 |
ENST00000263798.8
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr11_-_11353241 | 0.42 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr9_+_75088498 | 0.41 |
ENST00000346234.7
|
OSTF1
|
osteoclast stimulating factor 1 |
| chr3_+_52211442 | 0.40 |
ENST00000459884.1
|
ALAS1
|
5'-aminolevulinate synthase 1 |
| chr1_-_32901330 | 0.35 |
ENST00000329151.5
ENST00000373463.8 |
TMEM54
|
transmembrane protein 54 |
| chr9_-_75088198 | 0.34 |
ENST00000376808.8
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr18_+_9474994 | 0.34 |
ENST00000019317.8
|
RALBP1
|
ralA binding protein 1 |
| chr19_+_10625507 | 0.33 |
ENST00000590857.5
ENST00000588688.5 ENST00000586078.5 ENST00000335757.10 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr2_+_167248638 | 0.33 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr4_+_155667096 | 0.33 |
ENST00000393832.7
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_155666718 | 0.31 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr18_+_9475494 | 0.30 |
ENST00000383432.8
|
RALBP1
|
ralA binding protein 1 |
| chr4_+_155666963 | 0.30 |
ENST00000455639.6
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr6_+_32153441 | 0.30 |
ENST00000414204.5
ENST00000361568.6 ENST00000395523.5 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr19_-_17245889 | 0.30 |
ENST00000291442.4
|
NR2F6
|
nuclear receptor subfamily 2 group F member 6 |
| chr7_-_139109337 | 0.29 |
ENST00000464606.5
|
ZC3HAV1
|
zinc finger CCCH-type containing, antiviral 1 |
| chr3_-_49104745 | 0.28 |
ENST00000635194.1
ENST00000306125.12 ENST00000634602.1 ENST00000414533.5 ENST00000635443.1 ENST00000452739.5 ENST00000635231.1 |
QARS1
|
glutaminyl-tRNA synthetase 1 |
| chr4_+_155667198 | 0.24 |
ENST00000296518.11
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr15_+_66386902 | 0.24 |
ENST00000307102.10
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr2_-_208254232 | 0.23 |
ENST00000415913.5
ENST00000415282.5 ENST00000446179.5 |
IDH1
|
isocitrate dehydrogenase (NADP(+)) 1 |
| chr1_+_43974902 | 0.22 |
ENST00000532642.5
ENST00000236067.8 ENST00000471859.6 ENST00000472174.7 |
ATP6V0B
|
ATPase H+ transporting V0 subunit b |
| chr12_-_3753091 | 0.22 |
ENST00000252322.1
ENST00000440314.7 |
CRACR2A
|
calcium release activated channel regulator 2A |
| chr19_+_57527316 | 0.22 |
ENST00000240719.7
ENST00000376233.8 ENST00000594943.1 ENST00000602149.1 |
ZNF549
|
zinc finger protein 549 |
| chr14_-_23071617 | 0.20 |
ENST00000357481.6
|
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr12_+_18261511 | 0.20 |
ENST00000538779.6
ENST00000433979.6 |
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr11_-_65780917 | 0.19 |
ENST00000532090.3
|
AP5B1
|
adaptor related protein complex 5 subunit beta 1 |
| chr4_+_155666827 | 0.19 |
ENST00000511507.5
ENST00000506455.6 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr6_-_31139063 | 0.17 |
ENST00000259845.5
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr12_+_56104527 | 0.14 |
ENST00000552766.5
ENST00000303305.11 |
PA2G4
|
proliferation-associated 2G4 |
| chrX_-_120469282 | 0.14 |
ENST00000200639.9
ENST00000371335.4 |
LAMP2
|
lysosomal associated membrane protein 2 |
| chr5_+_134967901 | 0.13 |
ENST00000282611.8
|
CATSPER3
|
cation channel sperm associated 3 |
| chr1_+_156061142 | 0.12 |
ENST00000361084.10
|
RAB25
|
RAB25, member RAS oncogene family |
| chr11_-_120138104 | 0.11 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr1_-_16212598 | 0.11 |
ENST00000270747.8
|
ARHGEF19
|
Rho guanine nucleotide exchange factor 19 |
| chr18_-_37565765 | 0.11 |
ENST00000361795.9
ENST00000420428.7 |
CELF4
|
CUGBP Elav-like family member 4 |
| chr18_-_37565714 | 0.11 |
ENST00000591287.5
ENST00000601019.5 ENST00000601392.5 |
CELF4
|
CUGBP Elav-like family member 4 |
| chr18_-_37565628 | 0.10 |
ENST00000334919.9
ENST00000591282.5 ENST00000588597.5 |
CELF4
|
CUGBP Elav-like family member 4 |
| chrX_+_147911910 | 0.10 |
ENST00000370475.9
|
FMR1
|
FMRP translational regulator 1 |
| chr18_+_9475450 | 0.10 |
ENST00000585015.6
|
RALBP1
|
ralA binding protein 1 |
| chr10_-_50885619 | 0.09 |
ENST00000373997.8
|
A1CF
|
APOBEC1 complementation factor |
| chrX_+_147912039 | 0.09 |
ENST00000334557.10
ENST00000439526.6 |
FMR1
|
FMRP translational regulator 1 |
| chr4_-_128288791 | 0.07 |
ENST00000613358.4
ENST00000520121.6 |
PGRMC2
|
progesterone receptor membrane component 2 |
| chr6_-_43059367 | 0.07 |
ENST00000230413.9
ENST00000487429.1 ENST00000388752.8 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr10_-_50885656 | 0.05 |
ENST00000374001.6
ENST00000395489.6 ENST00000282641.6 ENST00000395495.5 ENST00000373995.7 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr1_+_94418435 | 0.05 |
ENST00000647998.2
|
ABCD3
|
ATP binding cassette subfamily D member 3 |
| chr7_-_124765753 | 0.04 |
ENST00000303921.3
|
GPR37
|
G protein-coupled receptor 37 |
| chr10_+_80079036 | 0.04 |
ENST00000372273.7
|
TMEM254
|
transmembrane protein 254 |
| chr1_-_36397880 | 0.03 |
ENST00000315732.3
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr22_+_18077976 | 0.02 |
ENST00000399744.8
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr16_+_2514253 | 0.01 |
ENST00000565223.1
|
ATP6V0C
|
ATPase H+ transporting V0 subunit c |
| chr18_-_57803307 | 0.01 |
ENST00000648908.2
|
ATP8B1
|
ATPase phospholipid transporting 8B1 |
| chr19_-_14835252 | 0.00 |
ENST00000641666.1
ENST00000642030.1 ENST00000642000.1 |
OR7C1
|
olfactory receptor family 7 subfamily C member 1 |
| chr14_-_23071538 | 0.00 |
ENST00000555566.1
ENST00000338631.10 ENST00000557515.5 ENST00000397341.7 |
ACIN1
|
apoptotic chromatin condensation inducer 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.3 | 0.8 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 1.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 0.5 | GO:0032824 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.6 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.5 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 2.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.8 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.1 | 0.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 1.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.8 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.2 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 1.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.4 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.5 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 1.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.2 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.1 | 0.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 1.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 2.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 0.8 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.8 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.3 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.1 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |