Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for GSC_GSC2
Z-value: 0.53
Transcription factors associated with GSC_GSC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
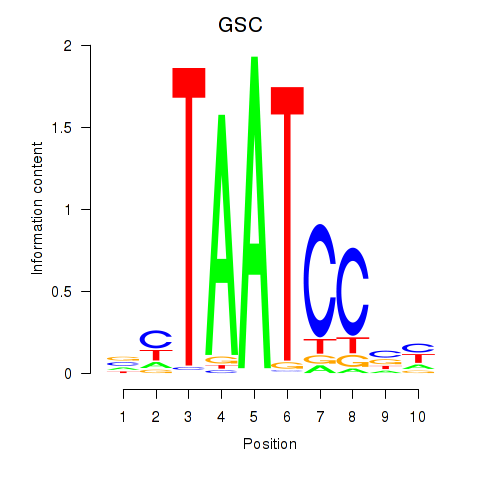
GSC
|
ENSG00000133937.5 | GSC |
|
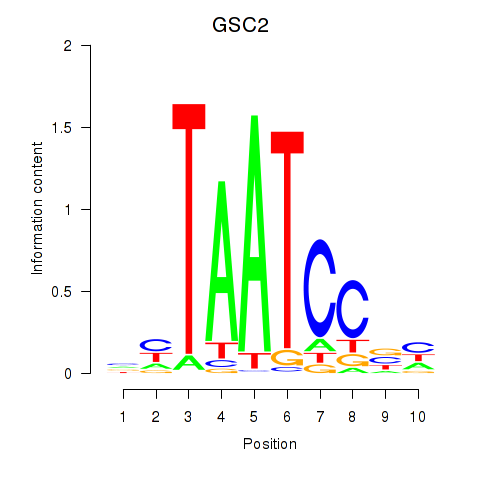
GSC2
|
ENSG00000063515.3 | GSC2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GSC2 | hg38_v1_chr22_-_19150292_19150292 | 0.37 | 4.5e-02 | Click! |
| GSC | hg38_v1_chr14_-_94770102_94770130 | -0.06 | 7.7e-01 | Click! |
Activity profile of GSC_GSC2 motif
Sorted Z-values of GSC_GSC2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GSC_GSC2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_40703744 | 1.58 |
ENST00000264651.3
|
KRT24
|
keratin 24 |
| chr19_-_51034892 | 1.41 |
ENST00000319590.8
ENST00000250351.4 |
KLK12
|
kallikrein related peptidase 12 |
| chr19_-_51034840 | 1.40 |
ENST00000529888.5
|
KLK12
|
kallikrein related peptidase 12 |
| chr22_-_19150292 | 1.22 |
ENST00000086933.3
|
GSC2
|
goosecoid homeobox 2 |
| chr19_-_51034993 | 1.16 |
ENST00000684732.1
|
KLK12
|
kallikrein related peptidase 12 |
| chr17_-_55511434 | 0.90 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36 |
| chr13_+_113667213 | 0.78 |
ENST00000335678.7
|
GRK1
|
G protein-coupled receptor kinase 1 |
| chr1_+_180928133 | 0.69 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr11_-_119346655 | 0.64 |
ENST00000360167.4
|
MFRP
|
membrane frizzled-related protein |
| chr5_+_38258373 | 0.52 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr11_-_111912871 | 0.47 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr6_-_35512863 | 0.47 |
ENST00000428978.1
ENST00000614066.4 ENST00000322263.8 |
TULP1
|
TUB like protein 1 |
| chr6_+_35342614 | 0.46 |
ENST00000337400.6
ENST00000311565.4 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr15_-_26939518 | 0.46 |
ENST00000541819.6
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3 |
| chr12_-_13095664 | 0.46 |
ENST00000337630.10
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr11_+_61755372 | 0.45 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor |
| chr19_-_48044037 | 0.43 |
ENST00000293255.3
|
CABP5
|
calcium binding protein 5 |
| chrX_+_136197020 | 0.42 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1 |
| chr17_-_50129792 | 0.41 |
ENST00000503131.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr12_-_1918473 | 0.40 |
ENST00000586184.5
ENST00000587995.5 ENST00000585732.1 |
CACNA2D4
|
calcium voltage-gated channel auxiliary subunit alpha2delta 4 |
| chr2_-_29074515 | 0.40 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr2_-_70553440 | 0.39 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr1_+_162381703 | 0.37 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr8_-_132111159 | 0.36 |
ENST00000673615.1
ENST00000434736.6 |
HHLA1
|
HERV-H LTR-associating 1 |
| chr10_+_123135938 | 0.35 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr11_-_111923722 | 0.35 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr1_+_202462730 | 0.35 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr19_+_47821907 | 0.32 |
ENST00000539067.5
ENST00000221996.12 ENST00000613299.1 |
CRX
|
cone-rod homeobox |
| chr16_+_81238682 | 0.32 |
ENST00000258168.7
ENST00000564552.1 |
BCO1
|
beta-carotene oxygenase 1 |
| chr19_-_49017087 | 0.30 |
ENST00000649238.3
|
LHB
|
luteinizing hormone subunit beta |
| chr7_-_128775793 | 0.29 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive |
| chr19_+_39196956 | 0.28 |
ENST00000339852.5
|
NCCRP1
|
NCCRP1, F-box associated domain containing |
| chr6_+_35342535 | 0.27 |
ENST00000360694.8
ENST00000418635.6 ENST00000448077.6 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr2_-_70553638 | 0.26 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr10_-_13972355 | 0.26 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr5_-_33984681 | 0.26 |
ENST00000296589.9
|
SLC45A2
|
solute carrier family 45 member 2 |
| chr11_-_62984957 | 0.26 |
ENST00000377871.7
ENST00000360421.9 |
SLC22A6
|
solute carrier family 22 member 6 |
| chr2_+_188291994 | 0.24 |
ENST00000409927.5
ENST00000409805.5 |
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr15_+_75647877 | 0.23 |
ENST00000308527.6
|
SNX33
|
sorting nexin 33 |
| chr2_-_174847015 | 0.23 |
ENST00000650938.1
|
CHN1
|
chimerin 1 |
| chr5_-_150758497 | 0.23 |
ENST00000521533.1
ENST00000424236.5 |
DCTN4
|
dynactin subunit 4 |
| chr5_+_150508110 | 0.22 |
ENST00000261797.7
|
NDST1
|
N-deacetylase and N-sulfotransferase 1 |
| chr15_+_58138368 | 0.22 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chrX_-_6228835 | 0.21 |
ENST00000381095.8
|
NLGN4X
|
neuroligin 4 X-linked |
| chrX_+_100644183 | 0.21 |
ENST00000640889.1
ENST00000373004.5 |
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr2_-_174846405 | 0.21 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr21_+_34668986 | 0.20 |
ENST00000349499.3
|
CLIC6
|
chloride intracellular channel 6 |
| chr2_-_29921580 | 0.20 |
ENST00000389048.8
|
ALK
|
ALK receptor tyrosine kinase |
| chr5_-_149944744 | 0.20 |
ENST00000255266.10
ENST00000617647.4 ENST00000613228.1 |
PDE6A
|
phosphodiesterase 6A |
| chr3_-_65597886 | 0.20 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr16_+_4371840 | 0.19 |
ENST00000304735.4
|
VASN
|
vasorin |
| chr7_-_92833896 | 0.19 |
ENST00000265734.8
|
CDK6
|
cyclin dependent kinase 6 |
| chr1_+_156147350 | 0.19 |
ENST00000435124.5
ENST00000633494.1 |
SEMA4A
|
semaphorin 4A |
| chr17_-_9791586 | 0.19 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chr3_+_4680617 | 0.18 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr14_-_100568070 | 0.18 |
ENST00000557378.6
ENST00000443071.6 ENST00000637646.1 |
BEGAIN
|
brain enriched guanylate kinase associated |
| chr1_+_159439722 | 0.18 |
ENST00000641630.1
ENST00000423932.6 |
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr16_+_57976435 | 0.18 |
ENST00000290871.10
ENST00000441824.4 |
TEPP
|
testis, prostate and placenta expressed |
| chr4_+_153222402 | 0.18 |
ENST00000676335.1
ENST00000675146.1 |
TRIM2
|
tripartite motif containing 2 |
| chr1_-_99766620 | 0.18 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr6_-_70283105 | 0.17 |
ENST00000683980.1
|
COL9A1
|
collagen type IX alpha 1 chain |
| chr2_-_55269038 | 0.17 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr10_-_102418748 | 0.17 |
ENST00000020673.6
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr6_-_70283184 | 0.17 |
ENST00000320755.12
|
COL9A1
|
collagen type IX alpha 1 chain |
| chr6_-_70283214 | 0.17 |
ENST00000683758.1
|
COL9A1
|
collagen type IX alpha 1 chain |
| chr22_-_30289607 | 0.16 |
ENST00000404953.7
|
CASTOR1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr14_-_100568475 | 0.16 |
ENST00000553553.6
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr15_-_43266857 | 0.16 |
ENST00000349114.8
ENST00000220420.10 |
TGM5
|
transglutaminase 5 |
| chr12_+_6840800 | 0.16 |
ENST00000541978.5
ENST00000229264.8 ENST00000435982.6 |
GNB3
|
G protein subunit beta 3 |
| chr3_-_193554952 | 0.16 |
ENST00000392443.7
|
ATP13A4
|
ATPase 13A4 |
| chr13_-_94479671 | 0.16 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr7_-_98252117 | 0.15 |
ENST00000420697.1
ENST00000415086.5 ENST00000447648.7 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr2_-_65432591 | 0.15 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr1_+_20070156 | 0.15 |
ENST00000375108.4
|
PLA2G5
|
phospholipase A2 group V |
| chr16_-_68236069 | 0.14 |
ENST00000473183.7
ENST00000565858.5 |
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr11_-_62984690 | 0.14 |
ENST00000421062.2
ENST00000458333.6 |
SLC22A6
|
solute carrier family 22 member 6 |
| chr4_+_153222307 | 0.14 |
ENST00000675899.1
ENST00000675611.1 ENST00000674872.1 ENST00000676167.1 |
TRIM2
|
tripartite motif containing 2 |
| chr11_-_70717994 | 0.14 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr17_-_9905248 | 0.13 |
ENST00000226193.6
|
RCVRN
|
recoverin |
| chr1_-_160343235 | 0.13 |
ENST00000368069.7
ENST00000241704.8 ENST00000647683.1 ENST00000649787.1 |
COPA
|
COPI coat complex subunit alpha |
| chr2_+_218607914 | 0.13 |
ENST00000417849.5
|
PLCD4
|
phospholipase C delta 4 |
| chr6_-_35512882 | 0.13 |
ENST00000229771.11
|
TULP1
|
TUB like protein 1 |
| chr3_-_54928044 | 0.13 |
ENST00000273286.6
|
LRTM1
|
leucine rich repeats and transmembrane domains 1 |
| chr7_+_29122274 | 0.13 |
ENST00000582692.2
ENST00000644824.1 |
ENSG00000285412.1
ENSG00000285162.1
|
novel transcript, antisense to CPVL chimerin 2 |
| chr11_-_63015831 | 0.12 |
ENST00000430500.6
ENST00000336232.7 |
SLC22A8
|
solute carrier family 22 member 8 |
| chr11_+_20022550 | 0.12 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr17_+_17042433 | 0.12 |
ENST00000651222.2
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr16_-_57971086 | 0.12 |
ENST00000564448.5
ENST00000311183.8 |
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr7_+_138797952 | 0.12 |
ENST00000397602.7
ENST00000442682.7 ENST00000458494.1 ENST00000413208.1 |
TMEM213
|
transmembrane protein 213 |
| chr15_+_86079863 | 0.11 |
ENST00000614907.3
ENST00000441037.7 |
AGBL1
|
ATP/GTP binding protein like 1 |
| chr14_-_21269392 | 0.11 |
ENST00000554891.5
ENST00000555883.5 ENST00000553753.5 ENST00000555914.5 ENST00000557336.1 ENST00000555215.5 ENST00000556628.5 ENST00000555137.5 ENST00000556226.5 ENST00000555309.5 ENST00000556142.5 ENST00000554969.5 ENST00000554455.5 ENST00000556513.5 ENST00000557201.5 ENST00000420743.6 ENST00000557768.1 ENST00000553300.6 ENST00000554383.5 ENST00000554539.5 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C |
| chr5_+_140401860 | 0.10 |
ENST00000532219.5
ENST00000394722.7 |
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr5_+_140401808 | 0.10 |
ENST00000616482.4
ENST00000297183.10 ENST00000360839.7 ENST00000421134.5 ENST00000394723.7 ENST00000511151.5 |
ANKHD1
|
ankyrin repeat and KH domain containing 1 |
| chr5_-_147081428 | 0.10 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr19_-_12723925 | 0.10 |
ENST00000425528.6
ENST00000589337.5 ENST00000588216.5 |
TNPO2
|
transportin 2 |
| chr11_-_67353503 | 0.10 |
ENST00000539074.1
ENST00000530584.5 ENST00000531239.2 ENST00000312419.8 ENST00000529704.5 |
POLD4
|
DNA polymerase delta 4, accessory subunit |
| chr5_-_115626161 | 0.10 |
ENST00000282382.8
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr6_-_127918604 | 0.10 |
ENST00000537166.5
|
THEMIS
|
thymocyte selection associated |
| chr3_-_197226351 | 0.10 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_172928165 | 0.10 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chrX_+_136196750 | 0.09 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1 |
| chr14_+_51860632 | 0.09 |
ENST00000555472.5
ENST00000556766.5 |
GNG2
|
G protein subunit gamma 2 |
| chrX_+_136197039 | 0.09 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chr1_-_27604135 | 0.08 |
ENST00000673934.1
ENST00000642245.1 |
AHDC1
|
AT-hook DNA binding motif containing 1 |
| chr1_-_27604176 | 0.08 |
ENST00000642416.1
|
AHDC1
|
AT-hook DNA binding motif containing 1 |
| chr20_-_50115935 | 0.08 |
ENST00000340309.7
ENST00000415862.6 ENST00000371677.7 |
UBE2V1
|
ubiquitin conjugating enzyme E2 V1 |
| chrX_-_139642518 | 0.07 |
ENST00000370573.8
ENST00000338585.6 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr5_-_147081462 | 0.07 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr12_-_52777343 | 0.07 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr5_-_33984636 | 0.07 |
ENST00000382102.7
ENST00000509381.1 |
SLC45A2
|
solute carrier family 45 member 2 |
| chr12_+_77830886 | 0.07 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chr7_-_105269007 | 0.07 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2 |
| chr19_-_49057730 | 0.07 |
ENST00000684222.1
ENST00000596965.5 |
CGB7
|
chorionic gonadotropin subunit beta 7 |
| chr6_-_41163182 | 0.07 |
ENST00000338469.3
|
TREM2
|
triggering receptor expressed on myeloid cells 2 |
| chr14_+_51860391 | 0.07 |
ENST00000335281.8
|
GNG2
|
G protein subunit gamma 2 |
| chr11_-_65857763 | 0.07 |
ENST00000531407.5
|
CFL1
|
cofilin 1 |
| chr10_+_100462969 | 0.06 |
ENST00000343737.6
|
WNT8B
|
Wnt family member 8B |
| chr4_+_169620527 | 0.06 |
ENST00000360642.7
ENST00000512813.5 ENST00000513761.6 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr17_+_40309161 | 0.06 |
ENST00000254066.10
|
RARA
|
retinoic acid receptor alpha |
| chr12_-_54385727 | 0.06 |
ENST00000551109.5
ENST00000546970.5 |
ZNF385A
|
zinc finger protein 385A |
| chrX_-_139642889 | 0.06 |
ENST00000370576.9
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr6_-_41163103 | 0.06 |
ENST00000373122.8
ENST00000373113.8 |
TREM2
|
triggering receptor expressed on myeloid cells 2 |
| chr11_-_82846128 | 0.06 |
ENST00000679809.1
ENST00000680186.1 ENST00000681592.1 |
PRCP
|
prolylcarboxypeptidase |
| chr17_+_4740005 | 0.06 |
ENST00000269289.10
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr6_-_65707214 | 0.06 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chrX_-_139642835 | 0.06 |
ENST00000536274.5
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr11_-_65857007 | 0.06 |
ENST00000527344.5
|
CFL1
|
cofilin 1 |
| chr1_+_18631006 | 0.05 |
ENST00000375375.7
|
PAX7
|
paired box 7 |
| chr17_-_4903088 | 0.05 |
ENST00000649488.2
|
CHRNE
|
cholinergic receptor nicotinic epsilon subunit |
| chr7_-_120857124 | 0.05 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr11_-_11353241 | 0.05 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr16_-_57971121 | 0.05 |
ENST00000251102.13
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chrX_-_124963768 | 0.05 |
ENST00000371130.7
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr2_+_218607861 | 0.05 |
ENST00000450993.7
|
PLCD4
|
phospholipase C delta 4 |
| chr11_-_82845734 | 0.05 |
ENST00000681883.1
ENST00000680040.1 ENST00000681432.1 |
PRCP
|
prolylcarboxypeptidase |
| chrX_-_110440218 | 0.05 |
ENST00000372057.1
ENST00000372054.3 |
AMMECR1
GNG5P2
|
AMMECR nuclear protein 1 G protein subunit gamma 5 pseudogene 2 |
| chr1_+_197268222 | 0.05 |
ENST00000367400.8
ENST00000638467.1 ENST00000367399.6 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr17_+_79024142 | 0.05 |
ENST00000579760.6
ENST00000339142.6 |
C1QTNF1
|
C1q and TNF related 1 |
| chr11_-_128842467 | 0.05 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr17_-_35560812 | 0.05 |
ENST00000674182.1
|
SLFN14
|
schlafen family member 14 |
| chr10_-_72954790 | 0.05 |
ENST00000373032.4
|
PLA2G12B
|
phospholipase A2 group XIIB |
| chr17_+_79024243 | 0.05 |
ENST00000311661.4
|
C1QTNF1
|
C1q and TNF related 1 |
| chr10_-_92243246 | 0.04 |
ENST00000412050.8
ENST00000614585.4 |
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr20_+_63865228 | 0.04 |
ENST00000611972.4
ENST00000615907.4 ENST00000369927.8 ENST00000346249.9 ENST00000348257.9 ENST00000352482.8 ENST00000351424.8 ENST00000217121.9 ENST00000358548.4 |
TPD52L2
|
TPD52 like 2 |
| chr9_-_35072561 | 0.04 |
ENST00000678650.1
|
VCP
|
valosin containing protein |
| chrX_+_66162663 | 0.04 |
ENST00000519389.6
|
HEPH
|
hephaestin |
| chr2_-_61017174 | 0.04 |
ENST00000407787.5
ENST00000398658.2 |
PUS10
|
pseudouridine synthase 10 |
| chr17_+_4739791 | 0.04 |
ENST00000433935.6
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr4_-_101347492 | 0.04 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr3_+_1092654 | 0.04 |
ENST00000350110.2
|
CNTN6
|
contactin 6 |
| chr1_+_165543992 | 0.04 |
ENST00000294818.2
|
LRRC52
|
leucine rich repeat containing 52 |
| chr17_-_42112674 | 0.04 |
ENST00000251642.8
ENST00000591220.5 |
DHX58
|
DExH-box helicase 58 |
| chr11_-_96343170 | 0.04 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr3_+_1093002 | 0.04 |
ENST00000446702.7
|
CNTN6
|
contactin 6 |
| chr17_-_28576882 | 0.04 |
ENST00000395319.7
ENST00000581807.5 ENST00000226253.9 ENST00000584086.5 ENST00000395321.6 |
ALDOC
|
aldolase, fructose-bisphosphate C |
| chr17_+_75525682 | 0.03 |
ENST00000392550.8
ENST00000167462.9 ENST00000375227.8 ENST00000578363.5 ENST00000579392.5 |
LLGL2
|
LLGL scribble cell polarity complex component 2 |
| chr1_+_113905156 | 0.03 |
ENST00000650596.1
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr22_+_22162155 | 0.03 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr19_-_39391029 | 0.03 |
ENST00000221265.8
|
PAF1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr22_+_29268316 | 0.03 |
ENST00000414183.6
ENST00000333395.10 ENST00000455726.5 ENST00000332035.10 |
EWSR1
|
EWS RNA binding protein 1 |
| chr5_-_150289941 | 0.03 |
ENST00000682786.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr8_+_69466617 | 0.03 |
ENST00000525061.5
ENST00000260128.8 ENST00000458141.6 |
SULF1
|
sulfatase 1 |
| chr22_-_38084093 | 0.03 |
ENST00000681075.1
|
SLC16A8
|
solute carrier family 16 member 8 |
| chr1_-_21279520 | 0.03 |
ENST00000357071.8
|
ECE1
|
endothelin converting enzyme 1 |
| chrX_+_12137409 | 0.03 |
ENST00000672010.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr1_-_31065671 | 0.03 |
ENST00000440538.6
ENST00000424085.6 ENST00000257075.9 ENST00000373747.7 ENST00000426105.7 ENST00000525843.5 ENST00000373742.6 |
PUM1
|
pumilio RNA binding family member 1 |
| chr18_-_26863187 | 0.03 |
ENST00000440832.7
|
AQP4
|
aquaporin 4 |
| chr2_-_55269207 | 0.02 |
ENST00000263629.9
ENST00000403721.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr17_+_4740042 | 0.02 |
ENST00000592813.5
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr17_+_49575828 | 0.02 |
ENST00000328741.6
|
NXPH3
|
neurexophilin 3 |
| chr4_+_70028452 | 0.02 |
ENST00000530128.5
ENST00000381057.3 ENST00000673563.1 |
HTN3
|
histatin 3 |
| chr3_-_49021045 | 0.02 |
ENST00000440857.5
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr7_-_78771108 | 0.02 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr20_-_41366805 | 0.02 |
ENST00000332312.4
|
EMILIN3
|
elastin microfibril interfacer 3 |
| chr11_-_67523396 | 0.02 |
ENST00000353903.9
ENST00000294288.5 |
CABP2
|
calcium binding protein 2 |
| chr9_+_100442271 | 0.02 |
ENST00000502978.1
|
MSANTD3-TMEFF1
|
MSANTD3-TMEFF1 readthrough |
| chr4_-_101347471 | 0.02 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr12_-_113136224 | 0.02 |
ENST00000546530.5
ENST00000261729.9 |
RASAL1
|
RAS protein activator like 1 |
| chr4_-_75902444 | 0.02 |
ENST00000286719.12
|
PPEF2
|
protein phosphatase with EF-hand domain 2 |
| chr10_+_102132994 | 0.02 |
ENST00000413464.6
ENST00000278070.7 |
PPRC1
|
PPARG related coactivator 1 |
| chr15_+_64460728 | 0.02 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr9_+_4839761 | 0.02 |
ENST00000448872.6
ENST00000441844.2 |
RCL1
|
RNA terminal phosphate cyclase like 1 |
| chr5_+_173889337 | 0.02 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chrX_-_24647300 | 0.02 |
ENST00000379144.7
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_-_113905020 | 0.02 |
ENST00000432415.5
ENST00000369571.2 ENST00000256658.8 ENST00000369564.5 |
AP4B1
|
adaptor related protein complex 4 subunit beta 1 |
| chr10_-_48274567 | 0.02 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr19_+_7914823 | 0.02 |
ENST00000565886.2
|
TGFBR3L
|
transforming growth factor beta receptor 3 like |
| chr5_-_16738341 | 0.02 |
ENST00000515803.5
|
MYO10
|
myosin X |
| chr18_-_37565628 | 0.02 |
ENST00000334919.9
ENST00000591282.5 ENST00000588597.5 |
CELF4
|
CUGBP Elav-like family member 4 |
| chr18_-_37565714 | 0.01 |
ENST00000591287.5
ENST00000601019.5 ENST00000601392.5 |
CELF4
|
CUGBP Elav-like family member 4 |
| chr15_+_64151706 | 0.01 |
ENST00000325881.9
|
SNX22
|
sorting nexin 22 |
| chr18_-_37565765 | 0.01 |
ENST00000361795.9
ENST00000420428.7 |
CELF4
|
CUGBP Elav-like family member 4 |
| chr17_+_46590669 | 0.01 |
ENST00000398238.8
|
NSF
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr2_+_127418420 | 0.01 |
ENST00000234071.8
ENST00000429925.5 ENST00000442644.5 |
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr3_-_10505508 | 0.01 |
ENST00000643662.1
ENST00000397077.6 ENST00000360273.7 |
ATP2B2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr6_-_37257622 | 0.01 |
ENST00000650812.1
ENST00000497775.1 ENST00000478262.2 ENST00000356757.7 |
ENSG00000286105.1
TMEM217
|
novel transmembrane protein transmembrane protein 217 |
| chr1_+_160343375 | 0.01 |
ENST00000294785.10
ENST00000421914.5 ENST00000438008.5 |
NCSTN
|
nicastrin |
| chr12_+_69739370 | 0.01 |
ENST00000550536.5
ENST00000362025.9 |
RAB3IP
|
RAB3A interacting protein |
| chrM_+_8489 | 0.01 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase membrane subunit 6 |
| chr10_-_125161019 | 0.01 |
ENST00000411419.6
|
CTBP2
|
C-terminal binding protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.3 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.1 | 0.4 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.2 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.2 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.1 | 0.7 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.2 | GO:0015855 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 5.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 1.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.1 | GO:2000814 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.0 | GO:1903004 | flavin adenine dinucleotide metabolic process(GO:0072387) regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.5 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 0.3 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.7 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |