Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
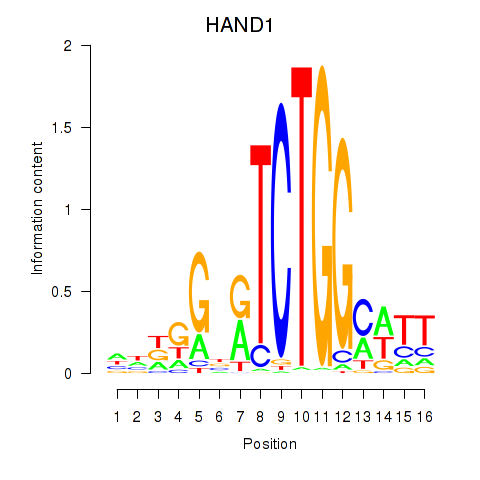
Results for HAND1
Z-value: 0.58
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.3 | HAND1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HAND1 | hg38_v1_chr5_-_154478218_154478234 | 0.23 | 2.2e-01 | Click! |
Activity profile of HAND1 motif
Sorted Z-values of HAND1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HAND1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_209579399 | 1.04 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr2_+_209579598 | 1.03 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr10_+_132036652 | 0.88 |
ENST00000657785.1
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr6_+_123804141 | 0.83 |
ENST00000368416.5
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr6_-_30684744 | 0.81 |
ENST00000615892.4
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr7_+_111091006 | 0.74 |
ENST00000451085.5
ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr7_+_111091119 | 0.73 |
ENST00000308478.10
|
LRRN3
|
leucine rich repeat neuronal 3 |
| chr11_-_102625332 | 0.68 |
ENST00000260228.3
|
MMP20
|
matrix metallopeptidase 20 |
| chr6_-_111873421 | 0.68 |
ENST00000368678.8
ENST00000523238.5 ENST00000354650.7 |
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr2_-_196926717 | 0.68 |
ENST00000409475.5
ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr20_-_57710539 | 0.66 |
ENST00000395816.7
ENST00000347215.8 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_+_227813834 | 0.64 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr2_-_27495185 | 0.62 |
ENST00000264703.4
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr1_+_65309517 | 0.61 |
ENST00000371069.5
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr9_-_136746006 | 0.58 |
ENST00000476567.1
|
LCN6
|
lipocalin 6 |
| chr2_+_209579429 | 0.55 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2 |
| chr11_+_118956289 | 0.54 |
ENST00000264031.3
|
UPK2
|
uroplakin 2 |
| chr22_+_24607638 | 0.53 |
ENST00000432867.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr10_+_132036336 | 0.53 |
ENST00000668452.1
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr2_+_113058637 | 0.53 |
ENST00000346807.7
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr2_+_203867943 | 0.50 |
ENST00000295854.10
ENST00000487393.1 ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr19_+_12751672 | 0.49 |
ENST00000549706.5
|
BEST2
|
bestrophin 2 |
| chr22_+_24607658 | 0.48 |
ENST00000451366.5
ENST00000428855.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr15_-_89679360 | 0.47 |
ENST00000300055.10
|
PLIN1
|
perilipin 1 |
| chr15_-_89679411 | 0.47 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr19_+_12751789 | 0.46 |
ENST00000553030.6
|
BEST2
|
bestrophin 2 |
| chr15_-_76012390 | 0.46 |
ENST00000394907.8
|
NRG4
|
neuregulin 4 |
| chr22_+_24607602 | 0.45 |
ENST00000447416.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_+_35868585 | 0.44 |
ENST00000652533.1
|
APLP1
|
amyloid beta precursor like protein 1 |
| chr10_-_116273606 | 0.44 |
ENST00000682743.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr19_+_35868518 | 0.43 |
ENST00000221891.9
|
APLP1
|
amyloid beta precursor like protein 1 |
| chr7_-_117323041 | 0.42 |
ENST00000491214.1
ENST00000265441.8 |
WNT2
|
Wnt family member 2 |
| chr12_+_49323236 | 0.40 |
ENST00000549275.5
ENST00000551245.5 ENST00000380327.9 ENST00000548311.5 ENST00000257909.8 ENST00000550346.5 ENST00000550709.5 ENST00000549534.1 ENST00000547807.5 ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr9_+_35042213 | 0.38 |
ENST00000378745.3
ENST00000312292.6 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr1_-_52404387 | 0.37 |
ENST00000371566.1
ENST00000371568.8 |
ORC1
|
origin recognition complex subunit 1 |
| chr12_+_40692413 | 0.37 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr20_+_46029165 | 0.37 |
ENST00000616201.4
ENST00000616202.4 ENST00000616933.4 ENST00000626937.2 |
SLC12A5
|
solute carrier family 12 member 5 |
| chr22_+_24603147 | 0.36 |
ENST00000412658.5
ENST00000445029.5 ENST00000400382.6 ENST00000419133.5 ENST00000438643.6 ENST00000452551.5 ENST00000412898.5 ENST00000400380.5 ENST00000455483.5 ENST00000430289.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr12_+_25958891 | 0.36 |
ENST00000381352.7
ENST00000535907.5 ENST00000405154.6 ENST00000615708.4 |
RASSF8
|
Ras association domain family member 8 |
| chr12_-_10802627 | 0.35 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7 |
| chr17_-_44947637 | 0.35 |
ENST00000587309.5
ENST00000593135.6 |
KIF18B
|
kinesin family member 18B |
| chr17_+_45844875 | 0.35 |
ENST00000329196.7
|
SPPL2C
|
signal peptide peptidase like 2C |
| chrX_+_50067576 | 0.32 |
ENST00000376108.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr19_+_507487 | 0.32 |
ENST00000359315.6
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr17_-_8210203 | 0.32 |
ENST00000578549.5
ENST00000582368.5 |
AURKB
|
aurora kinase B |
| chr17_-_76551174 | 0.30 |
ENST00000589145.1
|
CYGB
|
cytoglobin |
| chr2_-_207167220 | 0.30 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr11_+_125063295 | 0.30 |
ENST00000532000.5
ENST00000403796.7 ENST00000308074.4 |
SLC37A2
|
solute carrier family 37 member 2 |
| chr6_+_101181254 | 0.29 |
ENST00000682090.1
ENST00000421544.6 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr4_-_25030922 | 0.28 |
ENST00000382114.9
|
LGI2
|
leucine rich repeat LGI family member 2 |
| chr20_+_142573 | 0.27 |
ENST00000382398.4
|
DEFB126
|
defensin beta 126 |
| chr2_+_201132872 | 0.27 |
ENST00000470178.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr6_+_123803853 | 0.27 |
ENST00000368417.6
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr17_-_66220630 | 0.26 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr6_-_53148822 | 0.26 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr9_+_70043840 | 0.26 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2 |
| chr17_-_42028662 | 0.26 |
ENST00000461831.1
|
ZNF385C
|
zinc finger protein 385C |
| chr3_+_124094663 | 0.26 |
ENST00000460856.5
ENST00000240874.7 |
KALRN
|
kalirin RhoGEF kinase |
| chr11_-_2301859 | 0.25 |
ENST00000456145.2
ENST00000381153.8 |
C11orf21
|
chromosome 11 open reading frame 21 |
| chr8_-_92017292 | 0.25 |
ENST00000521553.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr5_-_147906530 | 0.25 |
ENST00000318315.5
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr6_-_87095059 | 0.24 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr17_+_39628496 | 0.24 |
ENST00000394265.5
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr2_+_203867764 | 0.23 |
ENST00000648405.2
|
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr1_-_155934398 | 0.22 |
ENST00000368320.7
ENST00000368321.8 |
KHDC4
|
KH domain containing 4, pre-mRNA splicing factor |
| chr12_-_53335737 | 0.22 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr6_+_43132472 | 0.22 |
ENST00000489707.5
|
PTK7
|
protein tyrosine kinase 7 (inactive) |
| chr11_-_19202004 | 0.22 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr16_+_6019585 | 0.21 |
ENST00000547372.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr12_-_53336342 | 0.21 |
ENST00000537210.2
ENST00000536324.4 |
SP7
|
Sp7 transcription factor |
| chr11_+_111540659 | 0.21 |
ENST00000375615.7
ENST00000525126.5 ENST00000375614.7 ENST00000533265.5 |
LAYN
|
layilin |
| chr19_-_12835400 | 0.20 |
ENST00000591512.2
ENST00000587549.1 ENST00000322912.9 |
RTBDN
|
retbindin |
| chr1_-_167914089 | 0.20 |
ENST00000476818.2
ENST00000367848.1 ENST00000367851.9 ENST00000545172.5 |
ADCY10
|
adenylate cyclase 10 |
| chr9_-_97938157 | 0.20 |
ENST00000616898.2
|
HEMGN
|
hemogen |
| chrX_+_150363258 | 0.20 |
ENST00000683696.1
|
MAMLD1
|
mastermind like domain containing 1 |
| chr19_-_12834548 | 0.19 |
ENST00000590404.6
ENST00000592204.5 ENST00000674343.2 |
RTBDN
|
retbindin |
| chr1_+_27392612 | 0.19 |
ENST00000374024.4
|
GPR3
|
G protein-coupled receptor 3 |
| chr18_+_12308232 | 0.19 |
ENST00000590103.5
ENST00000591909.5 ENST00000586653.5 ENST00000317702.10 ENST00000592683.5 ENST00000590967.5 ENST00000591208.1 ENST00000591463.1 |
TUBB6
|
tubulin beta 6 class V |
| chr1_+_11189347 | 0.19 |
ENST00000376819.4
|
ANGPTL7
|
angiopoietin like 7 |
| chr1_-_101996919 | 0.19 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chr10_+_103245887 | 0.18 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1 |
| chr9_-_120793377 | 0.18 |
ENST00000684001.1
ENST00000684405.1 ENST00000608872.6 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr19_-_37210484 | 0.18 |
ENST00000527838.5
ENST00000591492.5 ENST00000532828.7 |
ZNF585B
|
zinc finger protein 585B |
| chrX_+_150363306 | 0.18 |
ENST00000370401.7
ENST00000432680.7 |
MAMLD1
|
mastermind like domain containing 1 |
| chr13_-_60013178 | 0.18 |
ENST00000498416.2
ENST00000465066.5 |
DIAPH3
|
diaphanous related formin 3 |
| chrX_+_15749848 | 0.18 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chr11_+_2302119 | 0.17 |
ENST00000381121.7
|
TSPAN32
|
tetraspanin 32 |
| chr6_+_11093753 | 0.17 |
ENST00000416247.4
|
SMIM13
|
small integral membrane protein 13 |
| chr1_+_27935022 | 0.16 |
ENST00000411604.5
ENST00000373888.8 |
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr20_+_31368594 | 0.16 |
ENST00000253381.3
|
DEFB118
|
defensin beta 118 |
| chrX_+_71144818 | 0.16 |
ENST00000536169.5
ENST00000358741.4 ENST00000395855.6 ENST00000374051.7 |
NLGN3
|
neuroligin 3 |
| chr17_+_6444441 | 0.16 |
ENST00000250056.12
ENST00000571373.5 ENST00000570337.6 ENST00000572595.6 ENST00000572447.6 ENST00000576056.5 |
PIMREG
|
PICALM interacting mitotic regulator |
| chr10_+_50990864 | 0.16 |
ENST00000401604.8
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr2_+_201132769 | 0.15 |
ENST00000494258.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr1_+_43979179 | 0.15 |
ENST00000434555.7
ENST00000372324.6 ENST00000481924.2 |
B4GALT2
|
beta-1,4-galactosyltransferase 2 |
| chr17_-_42185452 | 0.15 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr4_-_103019634 | 0.15 |
ENST00000510559.1
ENST00000296422.12 ENST00000394789.7 |
SLC9B1
|
solute carrier family 9 member B1 |
| chr17_-_47831509 | 0.15 |
ENST00000414011.1
ENST00000351111.7 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr7_-_142885737 | 0.14 |
ENST00000359396.9
ENST00000436401.1 |
TRPV6
|
transient receptor potential cation channel subfamily V member 6 |
| chr3_+_141384790 | 0.14 |
ENST00000507722.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_+_116590956 | 0.14 |
ENST00000358834.9
ENST00000528052.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr10_+_116591010 | 0.13 |
ENST00000530319.5
ENST00000527980.5 ENST00000471549.5 ENST00000534537.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr17_+_38428456 | 0.13 |
ENST00000622683.5
ENST00000620417.4 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr9_-_120793343 | 0.13 |
ENST00000684047.1
|
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr2_+_201132928 | 0.13 |
ENST00000462763.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr11_-_26722051 | 0.11 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr12_-_52814106 | 0.11 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr19_-_12835421 | 0.11 |
ENST00000589272.5
|
RTBDN
|
retbindin |
| chr18_-_46756791 | 0.11 |
ENST00000538168.5
ENST00000536490.1 |
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr7_+_76510516 | 0.11 |
ENST00000257632.9
|
UPK3B
|
uroplakin 3B |
| chr14_+_96256194 | 0.11 |
ENST00000216629.11
ENST00000553356.1 |
BDKRB1
|
bradykinin receptor B1 |
| chr3_-_9769910 | 0.11 |
ENST00000256460.8
|
CAMK1
|
calcium/calmodulin dependent protein kinase I |
| chr15_-_31101707 | 0.10 |
ENST00000397795.6
ENST00000256552.11 ENST00000559179.2 |
TRPM1
|
transient receptor potential cation channel subfamily M member 1 |
| chr2_-_128027273 | 0.10 |
ENST00000259235.7
ENST00000357702.9 ENST00000424298.5 |
SAP130
|
Sin3A associated protein 130 |
| chr4_+_25160631 | 0.10 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chr1_+_207089195 | 0.10 |
ENST00000452902.6
|
C4BPB
|
complement component 4 binding protein beta |
| chr11_+_2301987 | 0.09 |
ENST00000612299.4
ENST00000182290.9 |
TSPAN32
|
tetraspanin 32 |
| chr5_+_144205250 | 0.09 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr5_-_181205182 | 0.09 |
ENST00000274773.12
|
TRIM7
|
tripartite motif containing 7 |
| chr3_+_148697784 | 0.09 |
ENST00000497524.5
ENST00000418473.7 ENST00000349243.8 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor type 1 |
| chr18_-_46757012 | 0.09 |
ENST00000315087.12
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr19_+_7920313 | 0.09 |
ENST00000221573.11
ENST00000595637.1 |
SNAPC2
|
small nuclear RNA activating complex polypeptide 2 |
| chr3_+_124094696 | 0.09 |
ENST00000360013.7
ENST00000684186.1 ENST00000684276.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr7_-_81770039 | 0.08 |
ENST00000222390.11
ENST00000453411.6 ENST00000457544.7 ENST00000444829.7 |
HGF
|
hepatocyte growth factor |
| chr21_+_33230073 | 0.08 |
ENST00000342101.7
ENST00000413881.5 ENST00000443073.5 |
IFNAR2
|
interferon alpha and beta receptor subunit 2 |
| chr7_+_97005538 | 0.08 |
ENST00000518156.3
|
DLX6
|
distal-less homeobox 6 |
| chr1_+_207089233 | 0.08 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr1_+_45012691 | 0.08 |
ENST00000469548.5
|
UROD
|
uroporphyrinogen decarboxylase |
| chr1_+_207089283 | 0.07 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr2_+_201132958 | 0.07 |
ENST00000479953.6
ENST00000340870.6 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr10_+_97584347 | 0.07 |
ENST00000370649.3
ENST00000370646.9 |
ENSG00000249967.1
HOGA1
|
novel protein 4-hydroxy-2-oxoglutarate aldolase 1 |
| chr1_+_45012896 | 0.07 |
ENST00000428106.1
|
UROD
|
uroporphyrinogen decarboxylase |
| chr1_+_207088825 | 0.06 |
ENST00000367078.8
|
C4BPB
|
complement component 4 binding protein beta |
| chr19_+_55488404 | 0.06 |
ENST00000594321.5
ENST00000389623.11 |
SSC5D
|
scavenger receptor cysteine rich family member with 5 domains |
| chr6_+_44223770 | 0.05 |
ENST00000652453.1
ENST00000393841.6 ENST00000371724.6 ENST00000642777.1 ENST00000645692.1 |
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr12_-_56360084 | 0.04 |
ENST00000314128.9
ENST00000557235.5 ENST00000651915.1 |
STAT2
|
signal transducer and activator of transcription 2 |
| chr2_-_42361198 | 0.04 |
ENST00000234301.3
|
COX7A2L
|
cytochrome c oxidase subunit 7A2 like |
| chr15_-_78811415 | 0.04 |
ENST00000388820.5
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif 7 |
| chr20_+_59721210 | 0.04 |
ENST00000395636.6
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr1_+_52404591 | 0.04 |
ENST00000257181.10
|
PRPF38A
|
pre-mRNA processing factor 38A |
| chr7_+_112450451 | 0.03 |
ENST00000429071.5
ENST00000403825.8 ENST00000675268.1 |
IFRD1
ENSG00000288634.1
|
interferon related developmental regulator 1 novel protein |
| chr14_+_21030509 | 0.03 |
ENST00000481535.5
|
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr12_+_109900258 | 0.03 |
ENST00000405876.9
|
TCHP
|
trichoplein keratin filament binding |
| chr12_+_67648737 | 0.03 |
ENST00000344096.4
ENST00000393555.3 |
DYRK2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr9_+_122614738 | 0.02 |
ENST00000297913.3
|
OR1Q1
|
olfactory receptor family 1 subfamily Q member 1 |
| chr3_+_112332500 | 0.02 |
ENST00000606471.5
|
CD200
|
CD200 molecule |
| chr11_+_67403887 | 0.02 |
ENST00000526387.5
|
TBC1D10C
|
TBC1 domain family member 10C |
| chr10_-_5003850 | 0.02 |
ENST00000421196.7
ENST00000455190.2 ENST00000380753.8 |
AKR1C2
|
aldo-keto reductase family 1 member C2 |
| chr17_+_7933080 | 0.02 |
ENST00000565740.5
|
CNTROB
|
centrobin, centriole duplication and spindle assembly protein |
| chr15_+_68277724 | 0.02 |
ENST00000306917.5
|
FEM1B
|
fem-1 homolog B |
| chr3_-_15341368 | 0.02 |
ENST00000408919.7
|
SH3BP5
|
SH3 domain binding protein 5 |
| chr14_+_73616844 | 0.02 |
ENST00000381139.1
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr11_+_67404077 | 0.02 |
ENST00000542590.2
ENST00000312390.9 |
TBC1D10C
|
TBC1 domain family member 10C |
| chr5_-_151087131 | 0.02 |
ENST00000315050.11
ENST00000523338.5 ENST00000522100.5 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr9_-_35828579 | 0.01 |
ENST00000377984.2
ENST00000423537.7 ENST00000472182.1 |
FAM221B
|
family with sequence similarity 221 member B |
| chr16_+_6019663 | 0.01 |
ENST00000422070.8
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr7_+_73827737 | 0.01 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr10_+_97584314 | 0.01 |
ENST00000370647.8
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr7_-_20217342 | 0.01 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr16_-_30031259 | 0.01 |
ENST00000380495.8
|
TLCD3B
|
TLC domain containing 3B |
| chr16_-_89490479 | 0.01 |
ENST00000642600.1
ENST00000301030.10 |
ANKRD11
|
ankyrin repeat domain 11 |
| chr7_-_151057848 | 0.00 |
ENST00000297518.4
|
CDK5
|
cyclin dependent kinase 5 |
| chr9_-_112332962 | 0.00 |
ENST00000458258.5
ENST00000210227.4 |
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr18_+_7567266 | 0.00 |
ENST00000580170.6
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.7 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 1.8 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.5 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.7 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.6 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.7 | GO:0070170 | enamel mineralization(GO:0070166) regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.3 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 2.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031696 | alpha-2C adrenergic receptor binding(GO:0031696) |
| 0.2 | 0.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.5 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 1.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 2.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.2 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |