Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
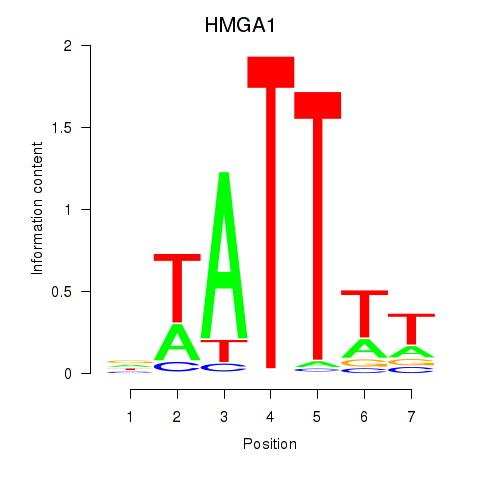
Results for HMGA1
Z-value: 0.92
Transcription factors associated with HMGA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA1
|
ENSG00000137309.20 | HMGA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA1 | hg38_v1_chr6_+_34236865_34236887 | 0.82 | 3.7e-08 | Click! |
Activity profile of HMGA1 motif
Sorted Z-values of HMGA1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153094521 | 3.25 |
ENST00000368750.8
|
SPRR2E
|
small proline rich protein 2E |
| chr9_-_41908681 | 3.12 |
ENST00000476961.5
|
CNTNAP3B
|
contactin associated protein family member 3B |
| chr16_-_46621345 | 3.09 |
ENST00000303383.8
|
SHCBP1
|
SHC binding and spindle associated 1 |
| chr3_-_74521140 | 3.00 |
ENST00000263665.6
|
CNTN3
|
contactin 3 |
| chr4_+_68447453 | 2.90 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr4_-_142305935 | 2.77 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr18_+_31447732 | 2.63 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr7_-_41703062 | 2.32 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr6_+_151325665 | 2.26 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr11_-_55936400 | 2.22 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr5_-_39270623 | 2.14 |
ENST00000512138.1
ENST00000646045.2 |
FYB1
|
FYN binding protein 1 |
| chr10_+_84452208 | 2.12 |
ENST00000480006.1
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr12_-_84892120 | 2.09 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr6_+_36029082 | 2.05 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr7_+_134745460 | 2.03 |
ENST00000436461.6
|
CALD1
|
caldesmon 1 |
| chr1_-_153070840 | 1.99 |
ENST00000368755.2
|
SPRR2B
|
small proline rich protein 2B |
| chr2_+_102311502 | 1.87 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr3_+_130931893 | 1.79 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr2_+_209580024 | 1.78 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr5_+_69167117 | 1.76 |
ENST00000506572.5
ENST00000256442.10 |
CCNB1
|
cyclin B1 |
| chr7_-_93890160 | 1.73 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr1_-_153041111 | 1.70 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr18_-_12656716 | 1.69 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr21_-_26843012 | 1.61 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 1.57 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr9_-_39288138 | 1.53 |
ENST00000297668.10
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr12_+_40692413 | 1.49 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr4_+_168092530 | 1.49 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr1_+_152908538 | 1.49 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr3_-_185821092 | 1.47 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr4_-_142305826 | 1.44 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr4_+_73436244 | 1.41 |
ENST00000226359.2
|
AFP
|
alpha fetoprotein |
| chr4_+_73436198 | 1.39 |
ENST00000395792.7
|
AFP
|
alpha fetoprotein |
| chr3_-_197260369 | 1.37 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr4_-_56681288 | 1.37 |
ENST00000556376.6
ENST00000420433.6 |
HOPX
|
HOP homeobox |
| chr10_-_104338431 | 1.33 |
ENST00000647721.1
ENST00000337478.3 |
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr2_+_190927649 | 1.30 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr3_-_197184131 | 1.30 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr10_+_116427839 | 1.30 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr15_-_99249523 | 1.29 |
ENST00000560235.1
ENST00000394132.7 ENST00000560860.5 ENST00000558078.5 ENST00000560772.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr16_-_18876305 | 1.28 |
ENST00000563235.5
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr17_-_59151794 | 1.28 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr12_-_50025394 | 1.27 |
ENST00000454520.6
ENST00000546595.5 ENST00000548824.5 ENST00000549777.5 ENST00000546723.5 ENST00000427314.6 ENST00000552157.5 ENST00000552310.5 ENST00000548644.5 ENST00000546786.5 ENST00000550149.5 ENST00000546764.5 ENST00000552004.2 ENST00000548320.5 ENST00000312377.10 ENST00000547905.5 ENST00000550651.5 ENST00000551145.5 ENST00000552921.5 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr11_-_7796942 | 1.26 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2 |
| chr20_+_3786772 | 1.26 |
ENST00000344256.10
ENST00000379598.9 |
CDC25B
|
cell division cycle 25B |
| chr4_+_154563003 | 1.25 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr5_-_147831627 | 1.24 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor Kazal type 1 |
| chr9_+_74615582 | 1.23 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr14_+_32329341 | 1.22 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr1_+_84164962 | 1.20 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr12_-_10826358 | 1.18 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chrX_+_80420466 | 1.17 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr6_+_27138588 | 1.15 |
ENST00000615353.1
|
H4C9
|
H4 clustered histone 9 |
| chr13_+_108596152 | 1.11 |
ENST00000356711.7
ENST00000251041.10 |
MYO16
|
myosin XVI |
| chr4_-_155866277 | 1.10 |
ENST00000537611.3
|
ASIC5
|
acid sensing ion channel subunit family member 5 |
| chr2_+_101697699 | 1.10 |
ENST00000350878.8
ENST00000350198.8 ENST00000324219.8 ENST00000425019.5 |
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_+_99638475 | 1.05 |
ENST00000452013.5
ENST00000261037.7 ENST00000652472.1 ENST00000273342.8 ENST00000621757.1 |
COL8A1
|
collagen type VIII alpha 1 chain |
| chr3_+_172754457 | 1.05 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr6_-_130956371 | 1.04 |
ENST00000639623.1
ENST00000525193.5 ENST00000527659.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr5_-_147831663 | 1.04 |
ENST00000296695.10
|
SPINK1
|
serine peptidase inhibitor Kazal type 1 |
| chr4_-_22443110 | 1.03 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr13_+_31945826 | 1.03 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr12_+_21527017 | 1.02 |
ENST00000535033.5
|
SPX
|
spexin hormone |
| chr1_-_158554405 | 1.02 |
ENST00000641282.1
ENST00000641622.1 |
OR6Y1
|
olfactory receptor family 6 subfamily Y member 1 |
| chr3_+_57890011 | 1.01 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr7_-_122699108 | 1.01 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr14_-_34713759 | 1.01 |
ENST00000673315.1
|
CFL2
|
cofilin 2 |
| chr10_+_74176537 | 1.00 |
ENST00000672394.1
|
ADK
|
adenosine kinase |
| chr3_-_122383218 | 0.99 |
ENST00000479899.5
ENST00000291458.9 ENST00000497726.5 |
MIX23
|
mitochondrial matrix import factor 23 |
| chr3_-_197183963 | 0.99 |
ENST00000653795.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr12_-_119804472 | 0.97 |
ENST00000678087.1
ENST00000677993.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr12_-_8650529 | 0.97 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr11_+_69641146 | 0.95 |
ENST00000227507.3
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr1_+_115029823 | 0.94 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr2_+_230759918 | 0.94 |
ENST00000614925.1
|
CAB39
|
calcium binding protein 39 |
| chr12_-_91111460 | 0.94 |
ENST00000266718.5
|
LUM
|
lumican |
| chr22_-_28306645 | 0.93 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr1_-_197067234 | 0.92 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr17_-_445939 | 0.92 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr3_-_149377637 | 0.92 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr1_-_28058087 | 0.91 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr1_+_192158448 | 0.91 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr8_-_121641424 | 0.91 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr11_+_89924064 | 0.90 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr7_+_130207847 | 0.90 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr5_-_126595237 | 0.89 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr22_-_28711931 | 0.89 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr6_-_25830557 | 0.89 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr18_+_63775395 | 0.87 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr19_-_48513161 | 0.87 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr3_-_197183849 | 0.87 |
ENST00000443183.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr17_-_41065879 | 0.86 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr15_+_80441229 | 0.86 |
ENST00000533983.5
ENST00000527771.5 ENST00000525103.1 |
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr9_+_72628020 | 0.85 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr11_+_55811367 | 0.84 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr3_-_151329539 | 0.84 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr10_-_93482194 | 0.84 |
ENST00000358334.9
ENST00000371488.3 |
MYOF
|
myoferlin |
| chr4_+_141636563 | 0.82 |
ENST00000320650.9
ENST00000296545.11 |
IL15
|
interleukin 15 |
| chr8_-_48921419 | 0.82 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr12_+_12891554 | 0.82 |
ENST00000014914.6
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A |
| chr9_-_39239174 | 0.81 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr4_+_141636611 | 0.80 |
ENST00000514653.5
|
IL15
|
interleukin 15 |
| chr2_-_162536955 | 0.80 |
ENST00000621889.1
|
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr1_+_84181630 | 0.80 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr12_-_16600703 | 0.80 |
ENST00000616247.4
|
LMO3
|
LIM domain only 3 |
| chr7_+_77840122 | 0.80 |
ENST00000450574.5
ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr2_+_209653171 | 0.79 |
ENST00000447185.5
|
MAP2
|
microtubule associated protein 2 |
| chr8_+_127737610 | 0.79 |
ENST00000652288.1
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr4_-_73988179 | 0.79 |
ENST00000296028.4
|
PPBP
|
pro-platelet basic protein |
| chr8_-_65789084 | 0.79 |
ENST00000379419.8
|
PDE7A
|
phosphodiesterase 7A |
| chr4_+_141636923 | 0.79 |
ENST00000529613.5
|
IL15
|
interleukin 15 |
| chr22_-_28712136 | 0.78 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2 |
| chr4_-_76007501 | 0.77 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr12_+_4809176 | 0.76 |
ENST00000280684.3
|
KCNA6
|
potassium voltage-gated channel subfamily A member 6 |
| chr19_+_926001 | 0.75 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr17_-_40984297 | 0.75 |
ENST00000377755.9
|
KRT40
|
keratin 40 |
| chr4_-_89835617 | 0.73 |
ENST00000611107.1
ENST00000345009.8 ENST00000505199.5 ENST00000502987.5 |
SNCA
|
synuclein alpha |
| chr4_-_39032343 | 0.72 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr7_-_92134737 | 0.71 |
ENST00000450723.5
|
CYP51A1
|
cytochrome P450 family 51 subfamily A member 1 |
| chr9_+_128340474 | 0.71 |
ENST00000300456.5
|
SLC27A4
|
solute carrier family 27 member 4 |
| chr8_-_61646807 | 0.70 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr1_+_108876973 | 0.70 |
ENST00000441735.2
ENST00000435987.5 ENST00000642355.1 ENST00000645164.2 ENST00000264126.9 ENST00000674914.1 ENST00000643094.1 ENST00000674700.1 |
GPSM2
|
G protein signaling modulator 2 |
| chr7_+_93921720 | 0.69 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr3_+_101827982 | 0.69 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr1_+_154429315 | 0.69 |
ENST00000476006.5
|
IL6R
|
interleukin 6 receptor |
| chr12_-_54259531 | 0.69 |
ENST00000550411.5
ENST00000439541.6 |
CBX5
|
chromobox 5 |
| chr8_+_12104389 | 0.69 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr12_-_86256267 | 0.69 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chrX_-_101617921 | 0.69 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr11_-_105139750 | 0.68 |
ENST00000530950.2
|
CARD18
|
caspase recruitment domain family member 18 |
| chr7_+_134866831 | 0.67 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr4_-_159035226 | 0.66 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr2_-_24328113 | 0.66 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr1_-_99766620 | 0.66 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr8_+_11809135 | 0.65 |
ENST00000528643.5
ENST00000525777.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr6_-_116829037 | 0.64 |
ENST00000368549.7
ENST00000530250.1 ENST00000310357.8 |
GPRC6A
|
G protein-coupled receptor class C group 6 member A |
| chr10_+_88594746 | 0.64 |
ENST00000531458.1
|
LIPJ
|
lipase family member J |
| chr1_+_178341445 | 0.64 |
ENST00000462775.5
|
RASAL2
|
RAS protein activator like 2 |
| chr12_+_96912517 | 0.64 |
ENST00000457368.2
|
NEDD1
|
NEDD1 gamma-tubulin ring complex targeting factor |
| chr6_+_116399395 | 0.64 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr12_-_21910853 | 0.64 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr3_-_51779187 | 0.64 |
ENST00000398780.5
ENST00000668964.1 ENST00000667863.2 ENST00000647442.1 |
IQCF6
|
IQ motif containing F6 |
| chr3_-_197183806 | 0.63 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr9_+_102995308 | 0.63 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr7_+_18495723 | 0.63 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr2_-_224947030 | 0.63 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr3_-_167474026 | 0.62 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr18_+_50375037 | 0.62 |
ENST00000398452.6
ENST00000417656.6 ENST00000488454.5 ENST00000285116.8 ENST00000494518.1 |
SKA1
|
spindle and kinetochore associated complex subunit 1 |
| chr11_+_5389377 | 0.61 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr10_-_93482326 | 0.61 |
ENST00000359263.9
|
MYOF
|
myoferlin |
| chr18_+_74499939 | 0.60 |
ENST00000584768.5
|
CNDP2
|
carnosine dipeptidase 2 |
| chr3_-_197226351 | 0.60 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chrX_-_132961390 | 0.60 |
ENST00000370836.6
ENST00000521489.5 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr10_+_119819244 | 0.60 |
ENST00000637174.1
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chrX_+_37780049 | 0.59 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr8_+_22995831 | 0.59 |
ENST00000522948.5
|
RHOBTB2
|
Rho related BTB domain containing 2 |
| chr9_+_94084458 | 0.59 |
ENST00000620992.5
ENST00000288976.3 |
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr13_-_46142834 | 0.59 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chrX_+_12906639 | 0.58 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8 |
| chr13_+_32031706 | 0.58 |
ENST00000542859.6
|
FRY
|
FRY microtubule binding protein |
| chr12_-_84911178 | 0.58 |
ENST00000681688.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr11_+_111255982 | 0.57 |
ENST00000637637.1
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr6_-_132659178 | 0.57 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr1_+_50103903 | 0.57 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr4_-_70666492 | 0.57 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr14_+_32329256 | 0.57 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr3_+_98166696 | 0.56 |
ENST00000641450.1
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr3_+_189631373 | 0.56 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr9_+_706841 | 0.56 |
ENST00000382293.7
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr5_+_55102635 | 0.56 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr2_-_210303608 | 0.54 |
ENST00000341685.8
|
MYL1
|
myosin light chain 1 |
| chr1_-_38876351 | 0.54 |
ENST00000360786.3
|
GJA9
|
gap junction protein alpha 9 |
| chr6_-_65707214 | 0.54 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chr20_+_31547367 | 0.54 |
ENST00000394552.3
|
MCTS2P
|
MCTS family member 2, pseudogene |
| chr11_+_111245725 | 0.54 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr5_+_137867852 | 0.53 |
ENST00000421631.6
ENST00000239926.9 |
MYOT
|
myotilin |
| chr6_-_32407123 | 0.53 |
ENST00000374993.4
ENST00000544175.2 ENST00000454136.7 ENST00000446536.2 |
BTNL2
|
butyrophilin like 2 |
| chr5_-_151924846 | 0.53 |
ENST00000274576.9
|
GLRA1
|
glycine receptor alpha 1 |
| chr8_+_38728550 | 0.53 |
ENST00000520340.5
ENST00000518415.5 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr14_-_106593319 | 0.52 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr5_-_126595185 | 0.52 |
ENST00000510111.6
ENST00000635851.1 ENST00000637964.1 ENST00000413020.6 ENST00000637782.1 ENST00000409134.8 ENST00000637272.1 ENST00000636879.1 ENST00000636743.1 ENST00000636886.1 ENST00000509270.2 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr8_-_94262308 | 0.52 |
ENST00000297596.3
ENST00000396194.6 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr12_+_71664352 | 0.51 |
ENST00000547843.1
|
THAP2
|
THAP domain containing 2 |
| chr9_-_21482313 | 0.51 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr9_-_92424427 | 0.51 |
ENST00000375550.5
|
OMD
|
osteomodulin |
| chr5_-_151924824 | 0.51 |
ENST00000455880.2
|
GLRA1
|
glycine receptor alpha 1 |
| chr17_-_82840010 | 0.51 |
ENST00000269394.4
ENST00000572562.1 |
ZNF750
|
zinc finger protein 750 |
| chr3_+_159069252 | 0.50 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr18_+_616672 | 0.50 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr17_-_66220630 | 0.50 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chrX_+_108045050 | 0.50 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr5_-_88731827 | 0.50 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr8_-_673547 | 0.50 |
ENST00000522893.1
|
ERICH1
|
glutamate rich 1 |
| chr1_+_67166448 | 0.49 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor |
| chr11_-_122116215 | 0.49 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr3_+_26694499 | 0.49 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chrX_+_41689006 | 0.49 |
ENST00000378138.5
ENST00000620846.1 ENST00000649219.1 |
GPR34
|
G protein-coupled receptor 34 |
| chr8_+_54616078 | 0.48 |
ENST00000220676.2
|
RP1
|
RP1 axonemal microtubule associated |
| chr2_+_135586250 | 0.48 |
ENST00000410054.5
ENST00000628915.2 |
R3HDM1
|
R3H domain containing 1 |
| chr9_-_42129125 | 0.48 |
ENST00000617422.4
ENST00000612828.4 ENST00000341990.8 ENST00000377561.7 ENST00000276974.7 |
CNTNAP3B
|
contactin associated protein family member 3B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 1.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.8 | 2.4 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.8 | 2.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.6 | 1.8 | GO:0060623 | regulation of chromosome condensation(GO:0060623) cellular response to iron(III) ion(GO:0071283) |
| 0.6 | 1.7 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.4 | 0.8 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.4 | 1.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.4 | 6.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.4 | 2.3 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.4 | 1.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.4 | 1.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 1.4 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.3 | 2.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 6.0 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.3 | 1.3 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 0.9 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.3 | 1.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 2.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 1.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 1.0 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.2 | 2.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.7 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 0.9 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.2 | 0.9 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 1.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.7 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 0.8 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.2 | 0.6 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 0.8 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 1.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.2 | 0.8 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 2.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 1.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 1.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 0.7 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 0.5 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 0.6 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 1.7 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.7 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.4 | GO:0072275 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.0 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 7.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.5 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.4 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.1 | 0.7 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 1.3 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.6 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.8 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.1 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) |
| 0.1 | 1.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.4 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.2 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 1.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 1.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 1.8 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.4 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.4 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.9 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.3 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.4 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 1.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.1 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 1.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.7 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.4 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.2 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.1 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 1.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.0 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.2 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.6 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 4.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.6 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.7 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 0.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.8 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 1.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 0.0 | 0.0 | GO:0045608 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 4.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.4 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.0 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 1.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 5.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 1.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0044791 | receptor-mediated endocytosis of virus by host cell(GO:0019065) modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 2.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.9 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 1.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.4 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.0 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.3 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 2.1 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 1.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.1 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.5 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.4 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.9 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 2.1 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 1.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 1.8 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 2.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 2.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.3 | 5.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 11.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 0.6 | GO:0071750 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.2 | 2.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 0.5 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 0.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 1.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 2.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 1.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.0 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 6.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.7 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 4.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.6 | 1.9 | GO:0042008 | interleukin-33 binding(GO:0002113) interleukin-18 receptor activity(GO:0042008) |
| 0.4 | 2.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.4 | 6.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 1.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 2.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 3.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.0 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.2 | 0.7 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 0.7 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 1.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 1.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 4.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.6 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 0.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 5.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 2.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.7 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 0.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 2.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.6 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 2.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.9 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.3 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 0.3 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.6 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 1.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 3.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.3 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 5.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 4.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 7.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 3.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 2.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 5.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 4.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 3.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 6.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 5.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.9 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |