Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HNF1A_HNF1B
Z-value: 0.94
Transcription factors associated with HNF1A_HNF1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
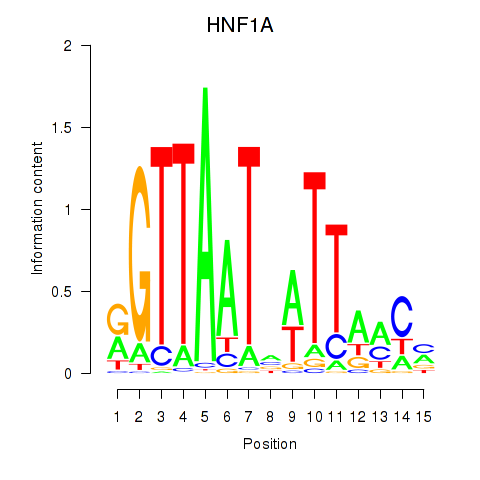
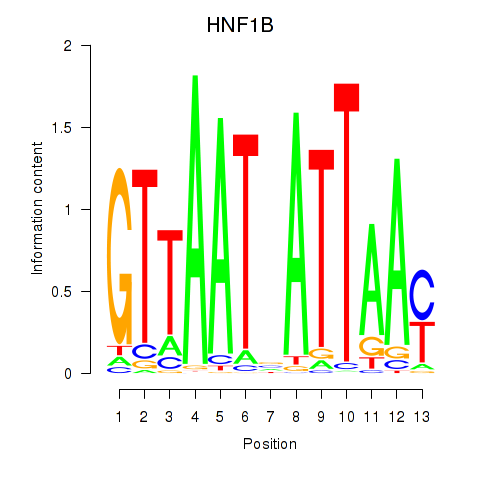
HNF1A
|
ENSG00000135100.19 | HNF1A |
|
HNF1B
|
ENSG00000275410.6 | HNF1B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF1A | hg38_v1_chr12_+_120978537_120978568 | -0.45 | 1.2e-02 | Click! |
| HNF1B | hg38_v1_chr17_-_37745018_37745069 | 0.16 | 3.9e-01 | Click! |
Activity profile of HNF1A_HNF1B motif
Sorted Z-values of HNF1A_HNF1B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF1A_HNF1B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_735026 | 7.37 |
ENST00000592155.5
ENST00000590161.2 |
PALM
|
paralemmin |
| chrX_-_15600953 | 3.31 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr4_-_109801978 | 2.60 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr1_+_103750406 | 2.30 |
ENST00000370079.3
|
AMY1C
|
amylase alpha 1C |
| chr1_+_103617427 | 2.29 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr19_+_49513353 | 2.28 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr1_+_18480930 | 2.26 |
ENST00000400664.3
|
KLHDC7A
|
kelch domain containing 7A |
| chr7_+_141995826 | 2.16 |
ENST00000549489.6
|
MGAM
|
maltase-glucoamylase |
| chr7_+_141995872 | 2.16 |
ENST00000497673.5
ENST00000620571.1 ENST00000475668.6 |
MGAM
|
maltase-glucoamylase |
| chr17_+_4788926 | 1.94 |
ENST00000331264.8
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr17_+_28473635 | 1.88 |
ENST00000314669.10
ENST00000545060.2 |
SLC13A2
|
solute carrier family 13 member 2 |
| chrX_-_15601077 | 1.85 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr18_+_58341038 | 1.83 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr17_+_28473278 | 1.73 |
ENST00000444914.7
|
SLC13A2
|
solute carrier family 13 member 2 |
| chr4_-_99352730 | 1.67 |
ENST00000510055.5
ENST00000515683.6 ENST00000511397.3 |
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide |
| chr16_+_20764036 | 1.64 |
ENST00000440284.6
|
ACSM3
|
acyl-CoA synthetase medium chain family member 3 |
| chr19_+_49513154 | 1.62 |
ENST00000426395.7
ENST00000600273.5 ENST00000599988.5 |
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr22_+_30607145 | 1.55 |
ENST00000405742.7
|
TCN2
|
transcobalamin 2 |
| chr1_+_162381703 | 1.54 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr22_+_30607167 | 1.50 |
ENST00000215838.8
|
TCN2
|
transcobalamin 2 |
| chr22_+_30607072 | 1.50 |
ENST00000450638.5
|
TCN2
|
transcobalamin 2 |
| chr8_-_94208548 | 1.41 |
ENST00000027335.8
ENST00000441892.6 ENST00000521491.1 |
CDH17
|
cadherin 17 |
| chr12_+_109347903 | 1.36 |
ENST00000310903.9
|
MYO1H
|
myosin IH |
| chr7_-_50565381 | 1.23 |
ENST00000444124.7
|
DDC
|
dopa decarboxylase |
| chr14_-_94388589 | 1.23 |
ENST00000402629.1
ENST00000556091.1 ENST00000393087.9 ENST00000554720.1 |
SERPINA1
|
serpin family A member 1 |
| chr6_-_29045175 | 1.20 |
ENST00000377175.2
|
OR2W1
|
olfactory receptor family 2 subfamily W member 1 |
| chr17_-_352784 | 1.17 |
ENST00000577079.5
ENST00000331302.12 ENST00000618002.4 ENST00000536489.6 |
RPH3AL
|
rabphilin 3A like (without C2 domains) |
| chr5_-_35195236 | 1.13 |
ENST00000509839.5
|
PRLR
|
prolactin receptor |
| chr17_+_70075215 | 1.11 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr17_+_70075317 | 1.11 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr6_-_154430495 | 1.03 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr22_-_32255344 | 0.98 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chrX_-_106038721 | 0.95 |
ENST00000372563.2
|
SERPINA7
|
serpin family A member 7 |
| chr3_-_138115594 | 0.89 |
ENST00000327532.7
ENST00000467030.5 |
DZIP1L
|
DAZ interacting zinc finger protein 1 like |
| chr3_-_157503574 | 0.88 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr13_-_103066411 | 0.85 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2 |
| chr16_+_21233672 | 0.85 |
ENST00000311620.7
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr9_-_121050264 | 0.82 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr17_-_28406160 | 0.79 |
ENST00000618626.1
ENST00000612814.5 |
SLC46A1
|
solute carrier family 46 member 1 |
| chr2_-_96505345 | 0.77 |
ENST00000310865.7
ENST00000451794.6 |
NEURL3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr4_-_1173168 | 0.71 |
ENST00000514490.5
ENST00000431380.5 ENST00000503765.5 |
SPON2
|
spondin 2 |
| chr13_+_95433593 | 0.70 |
ENST00000376873.7
|
CLDN10
|
claudin 10 |
| chr3_-_74521140 | 0.68 |
ENST00000263665.6
|
CNTN3
|
contactin 3 |
| chr2_+_37950432 | 0.64 |
ENST00000407257.5
ENST00000417700.6 ENST00000234195.7 ENST00000442857.5 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr5_+_141192330 | 0.63 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr16_-_55875299 | 0.63 |
ENST00000520435.2
|
CES5A
|
carboxylesterase 5A |
| chr6_-_52763473 | 0.62 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr6_-_25930611 | 0.62 |
ENST00000360488.7
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr12_-_39443390 | 0.62 |
ENST00000361961.7
|
KIF21A
|
kinesin family member 21A |
| chr4_-_69653223 | 0.62 |
ENST00000286604.8
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus |
| chr6_-_25930678 | 0.62 |
ENST00000377850.8
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr14_+_20688756 | 0.59 |
ENST00000397990.5
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin ribonuclease A family member 4 |
| chr16_-_55875358 | 0.59 |
ENST00000319165.13
ENST00000290567.14 |
CES5A
|
carboxylesterase 5A |
| chr4_-_99290975 | 0.59 |
ENST00000209668.3
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr2_+_132416795 | 0.57 |
ENST00000329321.4
|
GPR39
|
G protein-coupled receptor 39 |
| chr12_+_20810698 | 0.55 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr8_-_69833338 | 0.54 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr12_-_91180365 | 0.52 |
ENST00000547937.5
|
DCN
|
decorin |
| chr8_+_22053543 | 0.51 |
ENST00000519850.5
ENST00000381470.7 |
DMTN
|
dematin actin binding protein |
| chr2_-_227379297 | 0.51 |
ENST00000304568.4
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr22_-_30574572 | 0.51 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr6_+_125749623 | 0.51 |
ENST00000368364.4
|
HEY2
|
hes related family bHLH transcription factor with YRPW motif 2 |
| chr4_+_3441960 | 0.48 |
ENST00000382774.8
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr8_+_103880412 | 0.46 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_+_134237047 | 0.44 |
ENST00000370809.4
ENST00000517294.5 |
CCDC160
|
coiled-coil domain containing 160 |
| chr8_-_107498041 | 0.44 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1 |
| chr8_-_107497909 | 0.43 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1 |
| chr6_+_137871208 | 0.43 |
ENST00000614035.4
ENST00000621150.3 ENST00000619035.4 ENST00000615468.4 ENST00000620204.3 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr5_+_132369691 | 0.43 |
ENST00000245407.8
|
SLC22A5
|
solute carrier family 22 member 5 |
| chr9_+_128098304 | 0.41 |
ENST00000373064.9
|
SLC25A25
|
solute carrier family 25 member 25 |
| chr6_+_47656436 | 0.39 |
ENST00000507065.5
ENST00000296862.5 |
ADGRF2
|
adhesion G protein-coupled receptor F2 |
| chr5_+_68290637 | 0.38 |
ENST00000336483.9
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr6_+_54307856 | 0.37 |
ENST00000370869.7
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr16_-_69351778 | 0.37 |
ENST00000288025.4
|
TMED6
|
transmembrane p24 trafficking protein 6 |
| chr6_-_52087569 | 0.36 |
ENST00000340994.4
ENST00000371117.8 |
PKHD1
|
PKHD1 ciliary IPT domain containing fibrocystin/polyductin |
| chr9_+_79571767 | 0.34 |
ENST00000376544.7
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr9_+_79571956 | 0.34 |
ENST00000376552.8
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr20_+_44401397 | 0.33 |
ENST00000682427.1
ENST00000681977.1 ENST00000684136.1 ENST00000684046.1 ENST00000684476.1 ENST00000619550.5 ENST00000682169.1 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr11_-_65381951 | 0.33 |
ENST00000526432.5
ENST00000527174.5 |
SLC25A45
|
solute carrier family 25 member 45 |
| chr11_+_118533032 | 0.32 |
ENST00000526853.1
|
TMEM25
|
transmembrane protein 25 |
| chr3_-_49429252 | 0.32 |
ENST00000615713.4
|
NICN1
|
nicolin 1 |
| chr2_+_87748087 | 0.32 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr11_+_2384603 | 0.32 |
ENST00000527343.5
ENST00000464784.6 |
CD81
|
CD81 molecule |
| chr10_+_84173793 | 0.30 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr2_-_88128049 | 0.30 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chr6_+_131250375 | 0.30 |
ENST00000474850.2
|
AKAP7
|
A-kinase anchoring protein 7 |
| chr19_+_11239602 | 0.29 |
ENST00000252453.12
|
ANGPTL8
|
angiopoietin like 8 |
| chr8_-_17895487 | 0.29 |
ENST00000427924.5
ENST00000381841.4 |
FGL1
|
fibrinogen like 1 |
| chr3_-_161372821 | 0.28 |
ENST00000617024.1
ENST00000359175.8 |
SPTSSB
|
serine palmitoyltransferase small subunit B |
| chr11_-_117824734 | 0.27 |
ENST00000292079.7
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr12_+_25052732 | 0.27 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr11_+_560956 | 0.26 |
ENST00000397582.7
ENST00000397583.8 |
RASSF7
|
Ras association domain family member 7 |
| chr17_+_1742836 | 0.25 |
ENST00000324015.7
ENST00000450523.6 ENST00000453723.5 ENST00000453066.6 ENST00000382061.5 |
SERPINF2
|
serpin family F member 2 |
| chr20_+_44401222 | 0.25 |
ENST00000316099.9
|
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr12_-_8540873 | 0.25 |
ENST00000545274.5
ENST00000446457.6 ENST00000299663.8 |
CLEC4E
|
C-type lectin domain family 4 member E |
| chr17_+_37375974 | 0.25 |
ENST00000615133.2
ENST00000611038.4 |
C17orf78
|
chromosome 17 open reading frame 78 |
| chr1_+_207104226 | 0.25 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha |
| chr3_-_165078480 | 0.25 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr2_+_233617626 | 0.24 |
ENST00000373450.5
|
UGT1A8
|
UDP glucuronosyltransferase family 1 member A8 |
| chr4_+_119135825 | 0.24 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr12_+_100503352 | 0.24 |
ENST00000551379.5
ENST00000188403.7 |
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr6_+_54308429 | 0.23 |
ENST00000259782.9
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr12_+_100473708 | 0.23 |
ENST00000548884.5
ENST00000392986.8 |
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr6_-_160664270 | 0.22 |
ENST00000316300.10
|
LPA
|
lipoprotein(a) |
| chr1_+_119368773 | 0.22 |
ENST00000457318.5
ENST00000622548.4 ENST00000325945.4 |
HAO2
|
hydroxyacid oxidase 2 |
| chr12_+_100473951 | 0.22 |
ENST00000648861.1
ENST00000546380.1 |
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr4_+_99574812 | 0.21 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr11_+_46719193 | 0.21 |
ENST00000311907.10
ENST00000530231.5 ENST00000442468.1 |
F2
|
coagulation factor II, thrombin |
| chr19_+_1205761 | 0.21 |
ENST00000326873.12
ENST00000586243.5 |
STK11
|
serine/threonine kinase 11 |
| chr11_-_129024157 | 0.21 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr20_-_7940444 | 0.21 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr8_-_86743626 | 0.20 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3 |
| chr10_-_32378720 | 0.20 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1 |
| chr14_+_39233908 | 0.20 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr16_-_57797764 | 0.20 |
ENST00000465878.6
ENST00000561524.5 |
KIFC3
|
kinesin family member C3 |
| chr6_-_116060859 | 0.19 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr7_-_155809072 | 0.19 |
ENST00000430104.5
|
SHH
|
sonic hedgehog signaling molecule |
| chr1_-_42164737 | 0.19 |
ENST00000357001.3
|
GUCA2A
|
guanylate cyclase activator 2A |
| chr3_+_186666003 | 0.18 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein |
| chr10_-_126670686 | 0.18 |
ENST00000488181.3
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr16_+_14708944 | 0.18 |
ENST00000526520.5
ENST00000531598.6 |
NPIPA3
|
nuclear pore complex interacting protein family member A3 |
| chr15_+_86079863 | 0.18 |
ENST00000614907.3
ENST00000441037.7 |
AGBL1
|
ATP/GTP binding protein like 1 |
| chr2_-_162074182 | 0.18 |
ENST00000360534.8
|
DPP4
|
dipeptidyl peptidase 4 |
| chr10_+_110005804 | 0.17 |
ENST00000360162.7
|
ADD3
|
adducin 3 |
| chr7_+_152759744 | 0.17 |
ENST00000377776.7
ENST00000256001.13 ENST00000397282.2 |
ACTR3B
|
actin related protein 3B |
| chr13_+_33016415 | 0.17 |
ENST00000380099.4
|
KL
|
klotho |
| chr2_-_162074394 | 0.17 |
ENST00000676810.1
|
DPP4
|
dipeptidyl peptidase 4 |
| chr1_-_197067234 | 0.17 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr19_-_49061997 | 0.17 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr1_+_84301694 | 0.17 |
ENST00000394834.8
ENST00000370669.5 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr8_-_86230360 | 0.16 |
ENST00000419776.2
ENST00000297524.8 |
SLC7A13
|
solute carrier family 7 member 13 |
| chr3_+_184230373 | 0.15 |
ENST00000426955.6
|
VWA5B2
|
von Willebrand factor A domain containing 5B2 |
| chr9_+_137230757 | 0.15 |
ENST00000673865.1
ENST00000538474.5 ENST00000673835.1 ENST00000673953.1 ENST00000361134.2 |
SLC34A3
|
solute carrier family 34 member 3 |
| chr22_+_32043253 | 0.15 |
ENST00000266088.9
|
SLC5A1
|
solute carrier family 5 member 1 |
| chr6_-_25874212 | 0.15 |
ENST00000361703.10
ENST00000397060.8 |
SLC17A3
|
solute carrier family 17 member 3 |
| chr7_-_128910676 | 0.14 |
ENST00000620378.1
ENST00000610776.4 ENST00000613019.4 |
KCP
|
kielin cysteine rich BMP regulator |
| chr3_-_142963663 | 0.14 |
ENST00000340634.6
|
PAQR9
|
progestin and adipoQ receptor family member 9 |
| chr2_+_233760265 | 0.14 |
ENST00000305208.10
ENST00000360418.4 |
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1 |
| chr2_-_157444044 | 0.14 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr5_+_73813518 | 0.13 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr12_+_69739370 | 0.12 |
ENST00000550536.5
ENST00000362025.9 |
RAB3IP
|
RAB3A interacting protein |
| chr16_-_20576277 | 0.12 |
ENST00000566384.5
ENST00000565232.5 ENST00000567001.5 ENST00000565322.5 ENST00000569344.5 ENST00000329697.10 ENST00000568882.1 |
ACSM2B
|
acyl-CoA synthetase medium chain family member 2B |
| chr14_+_24120956 | 0.12 |
ENST00000558325.2
|
ENSG00000259371.2
|
novel protein |
| chr10_-_70888546 | 0.12 |
ENST00000299299.4
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase 1 |
| chr3_-_138174879 | 0.12 |
ENST00000260803.9
|
DBR1
|
debranching RNA lariats 1 |
| chr13_-_41019289 | 0.11 |
ENST00000239882.7
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr17_+_7407838 | 0.10 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr5_-_139383284 | 0.10 |
ENST00000353963.7
ENST00000348729.8 |
SLC23A1
|
solute carrier family 23 member 1 |
| chr14_-_57493827 | 0.10 |
ENST00000526336.1
ENST00000216445.8 |
CCDC198
|
coiled-coil domain containing 198 |
| chr7_-_88226987 | 0.10 |
ENST00000394641.7
|
SRI
|
sorcin |
| chr17_+_50746534 | 0.10 |
ENST00000511974.5
|
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr11_+_20599602 | 0.10 |
ENST00000525748.6
|
SLC6A5
|
solute carrier family 6 member 5 |
| chr16_+_20451596 | 0.10 |
ENST00000575690.5
ENST00000571894.1 |
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A |
| chr7_-_88226965 | 0.10 |
ENST00000490437.5
ENST00000431660.5 |
SRI
|
sorcin |
| chr15_-_55408018 | 0.10 |
ENST00000569205.5
|
CCPG1
|
cell cycle progression 1 |
| chr20_+_44401269 | 0.09 |
ENST00000443598.6
ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr10_-_37857582 | 0.09 |
ENST00000395867.8
ENST00000611278.4 |
ZNF248
|
zinc finger protein 248 |
| chr16_+_20451563 | 0.09 |
ENST00000417235.6
ENST00000219054.10 |
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A |
| chr22_-_38110675 | 0.09 |
ENST00000381669.8
|
BAIAP2L2
|
BAR/IMD domain containing adaptor protein 2 like 2 |
| chr16_+_81238682 | 0.08 |
ENST00000258168.7
ENST00000564552.1 |
BCO1
|
beta-carotene oxygenase 1 |
| chr3_+_180602156 | 0.08 |
ENST00000296015.9
ENST00000491380.5 ENST00000412756.6 ENST00000382584.8 |
TTC14
|
tetratricopeptide repeat domain 14 |
| chr13_+_27251569 | 0.08 |
ENST00000272274.8
ENST00000319826.8 ENST00000326092.8 |
RPL21
|
ribosomal protein L21 |
| chr7_+_33904911 | 0.08 |
ENST00000297161.6
|
BMPER
|
BMP binding endothelial regulator |
| chr11_+_64591214 | 0.08 |
ENST00000377574.6
ENST00000473690.5 |
SLC22A12
|
solute carrier family 22 member 12 |
| chr11_-_83682414 | 0.08 |
ENST00000404783.7
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr5_+_141392616 | 0.07 |
ENST00000398604.3
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr12_+_100473916 | 0.07 |
ENST00000549996.5
|
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr3_+_66220984 | 0.07 |
ENST00000354883.11
ENST00000336733.10 |
SLC25A26
|
solute carrier family 25 member 26 |
| chr12_-_23949642 | 0.07 |
ENST00000537393.5
ENST00000451604.7 ENST00000381381.6 |
SOX5
|
SRY-box transcription factor 5 |
| chr2_+_233729042 | 0.07 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr19_-_41627051 | 0.07 |
ENST00000221954.6
ENST00000600925.1 |
CEACAM4
|
CEA cell adhesion molecule 4 |
| chr7_-_44541262 | 0.06 |
ENST00000289547.8
ENST00000546276.5 ENST00000423141.1 |
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr9_-_91950201 | 0.06 |
ENST00000375708.4
|
ROR2
|
receptor tyrosine kinase like orphan receptor 2 |
| chr7_-_122304738 | 0.06 |
ENST00000442488.7
|
FEZF1
|
FEZ family zinc finger 1 |
| chr4_+_73404255 | 0.06 |
ENST00000621628.4
ENST00000621085.4 ENST00000415165.6 ENST00000295897.9 ENST00000503124.5 ENST00000509063.5 ENST00000401494.7 |
ALB
|
albumin |
| chr12_-_76486061 | 0.05 |
ENST00000548341.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr19_-_11529094 | 0.05 |
ENST00000588998.5
ENST00000586149.1 |
ECSIT
|
ECSIT signaling integrator |
| chr6_+_160702238 | 0.05 |
ENST00000366924.6
ENST00000308192.14 ENST00000418964.1 |
PLG
|
plasminogen |
| chr10_-_50885656 | 0.05 |
ENST00000374001.6
ENST00000395489.6 ENST00000282641.6 ENST00000395495.5 ENST00000373995.7 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr19_-_11529116 | 0.05 |
ENST00000592312.5
ENST00000590480.1 ENST00000585318.5 ENST00000270517.12 ENST00000252440.11 ENST00000417981.6 |
ECSIT
|
ECSIT signaling integrator |
| chr6_+_167122742 | 0.05 |
ENST00000341935.9
ENST00000349984.6 |
CCR6
|
C-C motif chemokine receptor 6 |
| chr17_+_50746614 | 0.05 |
ENST00000513969.5
ENST00000503728.1 |
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr3_+_107599309 | 0.04 |
ENST00000406780.5
|
BBX
|
BBX high mobility group box domain containing |
| chr1_+_207089283 | 0.04 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr1_+_207089233 | 0.03 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr10_-_50885619 | 0.03 |
ENST00000373997.8
|
A1CF
|
APOBEC1 complementation factor |
| chr4_+_154563003 | 0.03 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr2_+_233718734 | 0.03 |
ENST00000373409.8
|
UGT1A4
|
UDP glucuronosyltransferase family 1 member A4 |
| chr12_+_69738853 | 0.03 |
ENST00000247833.12
ENST00000378815.10 ENST00000483530.6 |
RAB3IP
|
RAB3A interacting protein |
| chr2_-_74328166 | 0.03 |
ENST00000358683.8
|
SLC4A5
|
solute carrier family 4 member 5 |
| chr6_+_25754699 | 0.03 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr2_-_162074050 | 0.03 |
ENST00000676768.1
|
DPP4
|
dipeptidyl peptidase 4 |
| chr16_+_20763990 | 0.03 |
ENST00000289416.10
|
ACSM3
|
acyl-CoA synthetase medium chain family member 3 |
| chr13_+_27251545 | 0.03 |
ENST00000311549.11
|
RPL21
|
ribosomal protein L21 |
| chr21_-_18403754 | 0.03 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15 |
| chr5_+_141182369 | 0.03 |
ENST00000609684.3
ENST00000625044.1 ENST00000623407.1 ENST00000623884.1 |
PCDHB16
ENSG00000279068.1
|
protocadherin beta 16 novel transcript |
| chr12_-_56221701 | 0.03 |
ENST00000615206.4
ENST00000549038.5 ENST00000552244.5 |
RNF41
|
ring finger protein 41 |
| chr2_+_233712905 | 0.02 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chrX_-_124963768 | 0.02 |
ENST00000371130.7
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr20_+_58839712 | 0.02 |
ENST00000313949.11
|
GNAS
|
GNAS complex locus |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.1 | 4.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 1.1 | 7.4 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.9 | 5.2 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.5 | 2.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.5 | 1.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.4 | 1.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.4 | 4.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.3 | 0.9 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.2 | 1.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 0.8 | GO:0001080 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.2 | 0.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 2.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.4 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.8 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.1 | 0.5 | GO:2000722 | negative regulation of vascular smooth muscle cell differentiation(GO:1905064) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) |
| 0.1 | 0.2 | GO:0060516 | primary prostatic bud elongation(GO:0060516) |
| 0.1 | 0.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.6 | GO:0032431 | diacylglycerol biosynthetic process(GO:0006651) activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 0.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.3 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.2 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 0.4 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.3 | GO:0051801 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) |
| 0.1 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.4 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.3 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 1.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) terpene metabolic process(GO:0042214) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 1.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 2.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 1.8 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 4.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 4.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.4 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 1.4 | 4.3 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 1.0 | 5.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.0 | 3.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.8 | 2.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.7 | 3.6 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 4.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 1.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 2.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 1.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 0.8 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.2 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.1 | 0.8 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.1 | 1.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 3.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 2.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |