Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
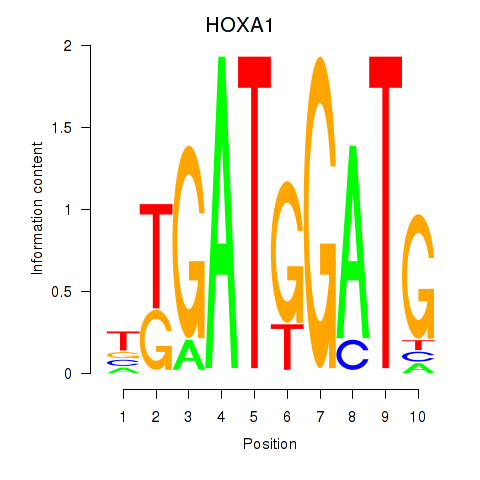
Results for HOXA1
Z-value: 0.74
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.10 | HOXA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA1 | hg38_v1_chr7_-_27095972_27096039 | -0.19 | 3.1e-01 | Click! |
Activity profile of HOXA1 motif
Sorted Z-values of HOXA1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_30470577 | 4.98 |
ENST00000435069.1
ENST00000540910.5 |
SEC14L3
|
SEC14 like lipid binding 3 |
| chr16_+_83968244 | 4.64 |
ENST00000305202.9
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr8_+_103140692 | 2.62 |
ENST00000438105.2
ENST00000309982.10 ENST00000297574.6 |
BAALC
|
BAALC binder of MAP3K1 and KLF4 |
| chrX_-_1212634 | 1.64 |
ENST00000381567.8
ENST00000381566.6 ENST00000400841.8 |
CRLF2
|
cytokine receptor like factor 2 |
| chr1_+_85062304 | 1.39 |
ENST00000326813.12
ENST00000528899.5 ENST00000294664.11 |
DNAI3
|
dynein axonemal intermediate chain 3 |
| chr3_-_112845950 | 1.36 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr15_-_93073111 | 1.24 |
ENST00000557420.1
ENST00000542321.6 |
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr3_+_98353854 | 1.13 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor family 5 subfamily K member 4 |
| chr10_-_27981805 | 0.98 |
ENST00000673512.1
ENST00000672877.1 ENST00000480504.1 |
ODAD2
|
outer dynein arm docking complex subunit 2 |
| chr14_+_96482982 | 0.96 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr8_-_27258414 | 0.95 |
ENST00000523048.5
|
STMN4
|
stathmin 4 |
| chr8_-_27611325 | 0.92 |
ENST00000523500.5
|
CLU
|
clusterin |
| chr15_+_81299416 | 0.91 |
ENST00000558332.3
|
IL16
|
interleukin 16 |
| chr8_-_27611424 | 0.85 |
ENST00000405140.7
|
CLU
|
clusterin |
| chr2_-_152098810 | 0.84 |
ENST00000636442.1
ENST00000638005.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chrX_+_135309480 | 0.80 |
ENST00000635820.1
|
ETDC
|
embryonic testis differentiation homolog C |
| chr1_+_18630839 | 0.78 |
ENST00000420770.7
|
PAX7
|
paired box 7 |
| chr17_+_70075215 | 0.76 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr13_+_35476740 | 0.74 |
ENST00000537702.5
|
NBEA
|
neurobeachin |
| chr13_-_35476682 | 0.71 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chr19_-_54242751 | 0.71 |
ENST00000245621.6
ENST00000396365.6 |
LILRA6
|
leukocyte immunoglobulin like receptor A6 |
| chr16_+_2988256 | 0.68 |
ENST00000573315.2
|
LINC00514
|
long intergenic non-protein coding RNA 514 |
| chr6_-_31684040 | 0.66 |
ENST00000375863.7
|
LY6G5C
|
lymphocyte antigen 6 family member G5C |
| chr9_+_36136416 | 0.66 |
ENST00000396613.7
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr4_-_137532452 | 0.65 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr9_-_35563867 | 0.64 |
ENST00000399742.7
ENST00000619051.4 |
FAM166B
|
family with sequence similarity 166 member B |
| chr8_-_27605271 | 0.63 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr22_+_25219633 | 0.62 |
ENST00000398215.3
|
CRYBB2
|
crystallin beta B2 |
| chr2_-_153478753 | 0.61 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog |
| chr1_+_3698027 | 0.60 |
ENST00000378290.4
|
TP73
|
tumor protein p73 |
| chr11_-_5227063 | 0.60 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr5_-_134367144 | 0.59 |
ENST00000265334.9
|
CDKL3
|
cyclin dependent kinase like 3 |
| chr16_-_67393486 | 0.57 |
ENST00000562206.1
ENST00000393957.7 ENST00000290942.9 |
TPPP3
|
tubulin polymerization promoting protein family member 3 |
| chr12_+_8123899 | 0.56 |
ENST00000641376.1
|
CLEC4A
|
C-type lectin domain family 4 member A |
| chrX_+_129779930 | 0.55 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr12_-_101210232 | 0.53 |
ENST00000536262.3
|
SLC5A8
|
solute carrier family 5 member 8 |
| chr6_+_25754699 | 0.53 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chrX_+_15507302 | 0.52 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr17_+_28473278 | 0.52 |
ENST00000444914.7
|
SLC13A2
|
solute carrier family 13 member 2 |
| chr10_-_87863533 | 0.49 |
ENST00000445946.5
|
KLLN
|
killin, p53 regulated DNA replication inhibitor |
| chr9_-_5304713 | 0.46 |
ENST00000381627.4
|
RLN2
|
relaxin 2 |
| chr11_+_125904467 | 0.46 |
ENST00000263576.11
ENST00000530414.5 ENST00000530129.6 |
DDX25
|
DEAD-box helicase 25 |
| chr20_-_31870629 | 0.45 |
ENST00000375966.8
ENST00000278979.7 |
DUSP15
|
dual specificity phosphatase 15 |
| chr7_-_117323041 | 0.45 |
ENST00000491214.1
ENST00000265441.8 |
WNT2
|
Wnt family member 2 |
| chr2_-_79086847 | 0.45 |
ENST00000454188.5
|
REG1B
|
regenerating family member 1 beta |
| chr11_+_34438900 | 0.44 |
ENST00000241052.5
|
CAT
|
catalase |
| chr9_-_10612966 | 0.43 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr20_-_25058115 | 0.43 |
ENST00000323482.9
|
ACSS1
|
acyl-CoA synthetase short chain family member 1 |
| chr10_+_7703340 | 0.42 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7703300 | 0.42 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_-_40294089 | 0.42 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr16_+_25216943 | 0.41 |
ENST00000219660.6
|
AQP8
|
aquaporin 8 |
| chr7_-_137343688 | 0.41 |
ENST00000348225.7
|
PTN
|
pleiotrophin |
| chr7_-_137343752 | 0.41 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr15_-_52295792 | 0.41 |
ENST00000261839.12
|
MYO5C
|
myosin VC |
| chr5_-_137735997 | 0.39 |
ENST00000505853.1
|
KLHL3
|
kelch like family member 3 |
| chr11_-_5441514 | 0.38 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1 |
| chr15_-_43220989 | 0.38 |
ENST00000540029.5
ENST00000441366.7 ENST00000648595.1 |
EPB42
|
erythrocyte membrane protein band 4.2 |
| chr13_+_111114550 | 0.38 |
ENST00000317133.9
|
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr9_+_2717479 | 0.37 |
ENST00000382082.4
|
KCNV2
|
potassium voltage-gated channel modifier subfamily V member 2 |
| chr7_+_120988683 | 0.36 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr19_-_49020523 | 0.35 |
ENST00000637680.1
|
ENSG00000268655.2
|
novel protein |
| chr17_+_62370218 | 0.35 |
ENST00000450662.7
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr12_+_75334675 | 0.34 |
ENST00000312442.2
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr1_-_91886144 | 0.33 |
ENST00000212355.9
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr1_+_65264694 | 0.33 |
ENST00000263441.11
ENST00000395325.7 |
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr22_-_38794111 | 0.33 |
ENST00000406622.5
ENST00000216068.9 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein axonemal light chain 4 |
| chr8_+_2045058 | 0.32 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr1_-_7853054 | 0.32 |
ENST00000361696.10
|
UTS2
|
urotensin 2 |
| chr1_-_182672232 | 0.32 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr12_+_75334655 | 0.32 |
ENST00000378695.9
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr21_+_31873010 | 0.31 |
ENST00000270112.7
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr5_+_134526100 | 0.30 |
ENST00000395003.5
|
JADE2
|
jade family PHD finger 2 |
| chr1_+_236394268 | 0.30 |
ENST00000334232.9
|
EDARADD
|
EDAR associated death domain |
| chr13_+_29428603 | 0.30 |
ENST00000380808.6
|
MTUS2
|
microtubule associated scaffold protein 2 |
| chr8_-_113436883 | 0.30 |
ENST00000455883.2
ENST00000297405.10 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr17_-_40665121 | 0.30 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr12_-_86256267 | 0.29 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr17_-_48545077 | 0.29 |
ENST00000330070.6
|
HOXB2
|
homeobox B2 |
| chr4_-_38804783 | 0.28 |
ENST00000308979.7
ENST00000505940.1 ENST00000515861.5 |
TLR1
|
toll like receptor 1 |
| chr1_-_13347134 | 0.28 |
ENST00000334600.7
|
PRAMEF14
|
PRAME family member 14 |
| chr11_-_104164361 | 0.28 |
ENST00000302251.9
|
PDGFD
|
platelet derived growth factor D |
| chr20_+_9514562 | 0.27 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5 |
| chr5_-_137736066 | 0.27 |
ENST00000309755.9
|
KLHL3
|
kelch like family member 3 |
| chr11_-_40293102 | 0.27 |
ENST00000527150.5
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr2_-_49154507 | 0.27 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor |
| chr17_-_63972918 | 0.26 |
ENST00000435607.3
|
SCN4A
|
sodium voltage-gated channel alpha subunit 4 |
| chr1_-_203229660 | 0.26 |
ENST00000255427.7
ENST00000367229.6 |
CHIT1
|
chitinase 1 |
| chr5_-_20575850 | 0.26 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr14_+_99684283 | 0.26 |
ENST00000261835.8
|
CYP46A1
|
cytochrome P450 family 46 subfamily A member 1 |
| chr14_-_80941719 | 0.26 |
ENST00000557411.5
ENST00000555265.6 |
CEP128
|
centrosomal protein 128 |
| chr1_-_110519175 | 0.25 |
ENST00000369771.4
|
KCNA10
|
potassium voltage-gated channel subfamily A member 10 |
| chr11_-_104164126 | 0.25 |
ENST00000393158.7
|
PDGFD
|
platelet derived growth factor D |
| chr2_-_49154433 | 0.25 |
ENST00000454032.5
ENST00000304421.8 |
FSHR
|
follicle stimulating hormone receptor |
| chr20_+_2692736 | 0.25 |
ENST00000380648.9
ENST00000497450.5 |
EBF4
|
EBF family member 4 |
| chr11_+_2461432 | 0.24 |
ENST00000335475.6
|
KCNQ1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr12_+_48183602 | 0.23 |
ENST00000316554.5
|
CCDC184
|
coiled-coil domain containing 184 |
| chr5_-_42825884 | 0.23 |
ENST00000506577.5
|
SELENOP
|
selenoprotein P |
| chr20_+_31870628 | 0.23 |
ENST00000375938.8
|
TTLL9
|
tubulin tyrosine ligase like 9 |
| chrX_-_43973382 | 0.23 |
ENST00000642620.1
ENST00000647044.1 |
NDP
|
norrin cystine knot growth factor NDP |
| chr9_-_21368057 | 0.23 |
ENST00000449498.2
|
IFNA13
|
interferon alpha 13 |
| chr1_-_100178215 | 0.23 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr9_+_61190003 | 0.23 |
ENST00000619167.1
|
SPATA31A7
|
SPATA31 subfamily A member 7 |
| chr22_-_19431692 | 0.22 |
ENST00000340170.8
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr16_+_28292485 | 0.22 |
ENST00000341901.5
|
SBK1
|
SH3 domain binding kinase 1 |
| chr17_-_44199834 | 0.22 |
ENST00000587097.6
|
ATXN7L3
|
ataxin 7 like 3 |
| chr6_-_15586006 | 0.22 |
ENST00000462989.6
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr5_+_134525649 | 0.22 |
ENST00000282605.8
ENST00000681547.2 ENST00000361895.6 ENST00000402835.5 |
JADE2
|
jade family PHD finger 2 |
| chr12_-_95996302 | 0.21 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr2_+_70978380 | 0.21 |
ENST00000272421.10
ENST00000441349.5 ENST00000457410.5 |
ANKRD53
|
ankyrin repeat domain 53 |
| chr3_+_72888031 | 0.21 |
ENST00000389617.9
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr11_+_31816266 | 0.20 |
ENST00000644607.1
ENST00000646221.1 ENST00000643671.1 ENST00000643931.1 ENST00000642614.1 ENST00000642818.1 ENST00000645848.1 ENST00000506388.2 ENST00000645824.1 ENST00000532942.5 |
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA novel protein |
| chr6_+_89562308 | 0.20 |
ENST00000522441.5
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr2_+_1484663 | 0.20 |
ENST00000446278.5
ENST00000469607.3 |
TPO
|
thyroid peroxidase |
| chr1_+_100345018 | 0.20 |
ENST00000635056.2
ENST00000647005.1 |
CDC14A
|
cell division cycle 14A |
| chr6_-_41154326 | 0.20 |
ENST00000426005.6
ENST00000437044.2 ENST00000373127.8 |
TREML1
|
triggering receptor expressed on myeloid cells like 1 |
| chr16_+_75222609 | 0.20 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr18_-_76495191 | 0.20 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr6_+_101393699 | 0.19 |
ENST00000369134.9
ENST00000684068.1 ENST00000683903.1 ENST00000681975.1 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr3_-_49929800 | 0.19 |
ENST00000455683.6
|
MON1A
|
MON1 homolog A, secretory trafficking associated |
| chr3_+_49021605 | 0.19 |
ENST00000451378.2
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr11_-_69819410 | 0.19 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr14_-_106235582 | 0.19 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr1_+_19882374 | 0.19 |
ENST00000375120.4
|
OTUD3
|
OTU deubiquitinase 3 |
| chr17_+_81528370 | 0.19 |
ENST00000417245.7
ENST00000334850.7 |
FSCN2
|
fascin actin-bundling protein 2, retinal |
| chr3_+_49022077 | 0.19 |
ENST00000326925.11
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr5_-_134367070 | 0.19 |
ENST00000521118.5
|
CDKL3
|
cyclin dependent kinase like 3 |
| chr2_-_60550900 | 0.19 |
ENST00000643222.1
ENST00000643459.1 ENST00000489516.7 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr1_+_43300971 | 0.19 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr14_-_20305932 | 0.19 |
ENST00000258821.8
ENST00000553828.1 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr11_-_18791768 | 0.18 |
ENST00000358540.7
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5 |
| chr6_+_13013513 | 0.18 |
ENST00000675203.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr12_+_124993633 | 0.18 |
ENST00000341446.9
ENST00000671775.2 |
BRI3BP
|
BRI3 binding protein |
| chr7_-_99735093 | 0.18 |
ENST00000611620.4
ENST00000620220.6 ENST00000336374.4 |
CYP3A7-CYP3A51P
CYP3A7
|
CYP3A7-CYP3A51P readthrough cytochrome P450 family 3 subfamily A member 7 |
| chr11_+_18266254 | 0.18 |
ENST00000532858.5
ENST00000649195.1 ENST00000356524.9 ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr5_+_140834230 | 0.18 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr11_+_63938971 | 0.18 |
ENST00000539656.5
ENST00000377793.9 |
NAA40
|
N-alpha-acetyltransferase 40, NatD catalytic subunit |
| chr19_-_10587219 | 0.18 |
ENST00000591240.5
ENST00000589684.5 ENST00000591676.1 ENST00000250244.11 ENST00000590923.5 |
AP1M2
|
adaptor related protein complex 1 subunit mu 2 |
| chr8_-_78805515 | 0.18 |
ENST00000379113.6
ENST00000541183.2 |
IL7
|
interleukin 7 |
| chr8_-_78805306 | 0.18 |
ENST00000639719.1
ENST00000263851.9 |
IL7
|
interleukin 7 |
| chr7_+_139829242 | 0.18 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr12_-_84912705 | 0.17 |
ENST00000679933.1
ENST00000680260.1 ENST00000551010.2 ENST00000679453.1 ENST00000681281.1 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr3_+_111542178 | 0.17 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr11_-_116772972 | 0.17 |
ENST00000375445.7
ENST00000260210.5 |
BUD13
|
BUD13 homolog |
| chr13_-_77919459 | 0.17 |
ENST00000643890.1
|
EDNRB
|
endothelin receptor type B |
| chr1_+_161153968 | 0.17 |
ENST00000368003.6
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr9_+_60914407 | 0.17 |
ENST00000437823.5
|
SPATA31A5
|
SPATA31 subfamily A member 5 |
| chrX_+_83508284 | 0.17 |
ENST00000644024.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr3_+_44874606 | 0.17 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr13_-_37598750 | 0.17 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr12_+_63779894 | 0.16 |
ENST00000261234.11
|
RXYLT1
|
ribitol xylosyltransferase 1 |
| chr17_-_28661897 | 0.16 |
ENST00000247020.9
|
SDF2
|
stromal cell derived factor 2 |
| chrX_+_8465426 | 0.16 |
ENST00000381029.4
|
VCX3B
|
variable charge X-linked 3B |
| chrM_+_9207 | 0.16 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chrX_-_10677720 | 0.16 |
ENST00000453318.6
|
MID1
|
midline 1 |
| chr2_+_74458400 | 0.16 |
ENST00000393972.7
ENST00000233615.7 ENST00000409737.5 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr3_+_133038366 | 0.16 |
ENST00000321871.11
ENST00000393130.7 ENST00000514894.5 ENST00000512662.5 |
TMEM108
|
transmembrane protein 108 |
| chrY_-_13986473 | 0.16 |
ENST00000250825.5
|
VCY
|
variable charge Y-linked |
| chr4_-_144140683 | 0.16 |
ENST00000324022.14
|
GYPA
|
glycophorin A (MNS blood group) |
| chr11_+_66480007 | 0.16 |
ENST00000531863.5
ENST00000532677.5 |
DPP3
|
dipeptidyl peptidase 3 |
| chr1_+_13171848 | 0.15 |
ENST00000415919.3
|
PRAMEF9
|
PRAME family member 9 |
| chr1_-_44355061 | 0.15 |
ENST00000649995.1
ENST00000457571.1 ENST00000372257.7 ENST00000452396.5 |
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr12_-_54984667 | 0.15 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr6_-_29005313 | 0.15 |
ENST00000377179.4
|
ZNF311
|
zinc finger protein 311 |
| chr17_-_27893339 | 0.15 |
ENST00000460380.6
ENST00000379102.8 ENST00000508862.5 ENST00000582441.1 |
LYRM9
ENSG00000266202.1
|
LYR motif containing 9 novel protein |
| chr8_+_42338477 | 0.15 |
ENST00000518925.5
ENST00000265421.9 |
POLB
|
DNA polymerase beta |
| chr16_+_21663968 | 0.15 |
ENST00000646100.2
|
OTOA
|
otoancorin |
| chr8_-_118111806 | 0.15 |
ENST00000378204.7
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr6_+_17393576 | 0.15 |
ENST00000229922.7
ENST00000611958.4 |
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr3_+_179604785 | 0.15 |
ENST00000611971.4
ENST00000493866.5 ENST00000259037.8 ENST00000472629.5 ENST00000482604.5 |
NDUFB5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr19_+_48756067 | 0.15 |
ENST00000222157.5
|
FGF21
|
fibroblast growth factor 21 |
| chr1_-_100895132 | 0.15 |
ENST00000535414.5
|
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr6_+_17393505 | 0.14 |
ENST00000616440.4
|
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr17_-_3292600 | 0.14 |
ENST00000615105.1
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1 |
| chr7_-_50093204 | 0.14 |
ENST00000419417.5
ENST00000046087.7 |
ZPBP
|
zona pellucida binding protein |
| chr12_-_88029385 | 0.14 |
ENST00000298699.7
ENST00000550553.5 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr13_+_113209597 | 0.14 |
ENST00000488558.2
ENST00000375440.9 |
CUL4A
|
cullin 4A |
| chr16_-_4752565 | 0.13 |
ENST00000588099.1
ENST00000588942.1 |
ENSG00000266994.1
ZNF500
|
novel transcript zinc finger protein 500 |
| chr6_+_32024278 | 0.13 |
ENST00000647698.1
|
C4B
|
complement C4B (Chido blood group) |
| chr16_+_30378312 | 0.13 |
ENST00000528032.5
ENST00000622647.3 |
ZNF48
|
zinc finger protein 48 |
| chr10_-_95693893 | 0.13 |
ENST00000371209.5
ENST00000680144.1 ENST00000430368.6 ENST00000371217.10 |
TCTN3
|
tectonic family member 3 |
| chr11_+_4704782 | 0.13 |
ENST00000380390.6
|
MMP26
|
matrix metallopeptidase 26 |
| chr16_-_88686453 | 0.13 |
ENST00000332281.6
|
SNAI3
|
snail family transcriptional repressor 3 |
| chr19_+_13764502 | 0.13 |
ENST00000040663.8
ENST00000319545.12 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr1_+_202203721 | 0.13 |
ENST00000255432.11
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6 |
| chr6_-_168319691 | 0.13 |
ENST00000610183.1
ENST00000607983.1 |
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr12_-_21774688 | 0.13 |
ENST00000240662.3
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr3_-_71064964 | 0.13 |
ENST00000650387.1
|
FOXP1
|
forkhead box P1 |
| chr2_+_64988469 | 0.13 |
ENST00000531327.5
|
SLC1A4
|
solute carrier family 1 member 4 |
| chr6_-_168319762 | 0.13 |
ENST00000366795.4
|
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr10_-_101839818 | 0.13 |
ENST00000348850.9
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr1_-_23217482 | 0.12 |
ENST00000374619.2
|
HTR1D
|
5-hydroxytryptamine receptor 1D |
| chrX_+_54808334 | 0.12 |
ENST00000218439.8
|
MAGED2
|
MAGE family member D2 |
| chr3_+_51829417 | 0.12 |
ENST00000440739.3
ENST00000444293.5 |
IQCF3
|
IQ motif containing F3 |
| chr4_-_112636858 | 0.12 |
ENST00000503172.5
ENST00000505019.6 ENST00000309071.9 |
ZGRF1
|
zinc finger GRF-type containing 1 |
| chr20_+_31879798 | 0.12 |
ENST00000620043.4
|
TTLL9
|
tubulin tyrosine ligase like 9 |
| chr3_-_150546403 | 0.12 |
ENST00000239944.7
ENST00000491660.1 ENST00000487153.1 |
SERP1
|
stress associated endoplasmic reticulum protein 1 |
| chr4_-_38782970 | 0.12 |
ENST00000502321.5
ENST00000308973.9 ENST00000613579.4 ENST00000361424.6 ENST00000622002.4 |
TLR10
|
toll like receptor 10 |
| chr5_-_151758639 | 0.12 |
ENST00000522710.1
|
ATOX1
|
antioxidant 1 copper chaperone |
| chr16_+_16379055 | 0.12 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family member A7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.5 | 2.4 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 0.8 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.3 | 0.8 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.2 | 0.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 0.5 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.2 | 0.7 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.2 | 0.5 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 0.4 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.4 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.3 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.3 | GO:0034699 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.3 | GO:1900108 | inner medullary collecting duct development(GO:0072061) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.2 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.2 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 1.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0045554 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.0 | 1.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.8 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.1 | GO:0032490 | detection of molecule of bacterial origin(GO:0032490) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.5 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.2 | 0.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.5 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 1.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 2.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 1.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 1.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |