Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
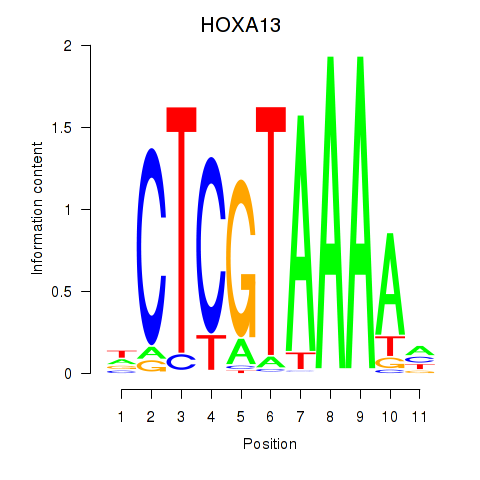
Results for HOXA13
Z-value: 0.49
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.9 | HOXA13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA13 | hg38_v1_chr7_-_27200083_27200096 | 0.02 | 9.2e-01 | Click! |
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_87407965 | 1.57 |
ENST00000369562.9
|
CFAP206
|
cilia and flagella associated protein 206 |
| chr10_-_59753388 | 0.98 |
ENST00000430431.5
|
MRLN
|
myoregulin |
| chr10_-_59753444 | 0.76 |
ENST00000594536.5
ENST00000414264.6 |
MRLN
|
myoregulin |
| chr7_-_16881967 | 0.66 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr10_-_27981805 | 0.61 |
ENST00000673512.1
ENST00000672877.1 ENST00000480504.1 |
ODAD2
|
outer dynein arm docking complex subunit 2 |
| chr7_+_23680130 | 0.57 |
ENST00000409192.7
ENST00000409653.5 ENST00000409994.3 ENST00000344962.9 |
FAM221A
|
family with sequence similarity 221 member A |
| chr3_-_197949869 | 0.49 |
ENST00000452735.1
ENST00000453254.5 ENST00000455191.5 |
IQCG
|
IQ motif containing G |
| chr16_+_82056423 | 0.45 |
ENST00000568090.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr15_+_70936487 | 0.43 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr11_+_101914997 | 0.42 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chr10_-_67838173 | 0.40 |
ENST00000225171.7
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr13_-_35855627 | 0.40 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr11_-_114595750 | 0.36 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chrX_-_38327496 | 0.36 |
ENST00000642395.2
ENST00000645032.1 ENST00000644238.1 ENST00000642558.1 ENST00000339363.7 ENST00000644337.1 ENST00000647261.1 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr12_+_20810698 | 0.36 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr11_-_114595777 | 0.35 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr1_+_47067212 | 0.34 |
ENST00000334194.4
|
CYP4Z1
|
cytochrome P450 family 4 subfamily Z member 1 |
| chr14_-_102240597 | 0.34 |
ENST00000523231.5
ENST00000524370.5 ENST00000517966.5 |
MOK
|
MOK protein kinase |
| chr12_+_45216014 | 0.32 |
ENST00000425752.6
|
ANO6
|
anoctamin 6 |
| chr14_-_91947654 | 0.30 |
ENST00000342058.9
|
FBLN5
|
fibulin 5 |
| chr3_+_129440196 | 0.30 |
ENST00000507564.5
ENST00000348417.7 ENST00000504021.5 ENST00000349441.6 |
IFT122
|
intraflagellar transport 122 |
| chr16_-_21278282 | 0.29 |
ENST00000572914.2
|
CRYM
|
crystallin mu |
| chr3_-_112846856 | 0.29 |
ENST00000488794.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr6_-_79537423 | 0.28 |
ENST00000369846.9
ENST00000392959.5 ENST00000467898.3 |
LCA5
|
lebercilin LCA5 |
| chr12_-_110920568 | 0.27 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr1_+_78303865 | 0.27 |
ENST00000370758.5
|
PTGFR
|
prostaglandin F receptor |
| chr4_-_169612571 | 0.25 |
ENST00000507142.6
ENST00000510533.5 ENST00000439128.6 ENST00000511633.5 ENST00000512193.5 |
NEK1
|
NIMA related kinase 1 |
| chr19_+_48993864 | 0.24 |
ENST00000595090.6
|
RUVBL2
|
RuvB like AAA ATPase 2 |
| chr13_-_35855758 | 0.24 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr14_-_106470788 | 0.24 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr13_+_23579346 | 0.23 |
ENST00000382258.8
ENST00000382263.3 |
TNFRSF19
|
TNF receptor superfamily member 19 |
| chr1_+_202462730 | 0.23 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr8_-_71356511 | 0.23 |
ENST00000419131.6
ENST00000388743.6 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_216805367 | 0.22 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr2_+_37344594 | 0.21 |
ENST00000404976.5
ENST00000338415.8 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr19_+_48993525 | 0.21 |
ENST00000601968.5
ENST00000596837.5 |
RUVBL2
|
RuvB like AAA ATPase 2 |
| chr1_-_111563934 | 0.21 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr8_-_71356653 | 0.21 |
ENST00000388742.8
ENST00000388740.4 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_-_100993448 | 0.21 |
ENST00000495063.6
ENST00000486770.7 ENST00000530539.2 |
ABI3BP
|
ABI family member 3 binding protein |
| chr3_-_179243284 | 0.20 |
ENST00000486944.2
|
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr5_+_141223332 | 0.20 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chrX_-_24672654 | 0.20 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr10_+_125896549 | 0.20 |
ENST00000368693.6
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr5_-_124745315 | 0.19 |
ENST00000306315.9
|
ZNF608
|
zinc finger protein 608 |
| chr3_-_160399207 | 0.18 |
ENST00000465537.5
ENST00000486856.5 ENST00000468218.5 ENST00000478370.5 ENST00000326448.12 |
IFT80
|
intraflagellar transport 80 |
| chr3_-_122793772 | 0.18 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr18_-_55587335 | 0.18 |
ENST00000638154.3
|
TCF4
|
transcription factor 4 |
| chr6_+_44342684 | 0.18 |
ENST00000288390.2
|
SPATS1
|
spermatogenesis associated serine rich 1 |
| chr14_+_22112280 | 0.18 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr2_+_108607140 | 0.18 |
ENST00000410093.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr9_-_39288138 | 0.17 |
ENST00000297668.10
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr18_-_27143024 | 0.17 |
ENST00000581714.5
|
CHST9
|
carbohydrate sulfotransferase 9 |
| chr6_+_44342639 | 0.17 |
ENST00000674044.1
ENST00000515220.5 ENST00000323108.12 |
SPATS1
|
spermatogenesis associated serine rich 1 |
| chr10_+_45000898 | 0.17 |
ENST00000298299.4
|
ZNF22
|
zinc finger protein 22 |
| chr3_+_4493340 | 0.16 |
ENST00000357086.10
ENST00000354582.12 ENST00000649015.2 ENST00000467056.6 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr3_-_114758940 | 0.16 |
ENST00000464560.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr22_+_18150162 | 0.16 |
ENST00000215794.8
|
USP18
|
ubiquitin specific peptidase 18 |
| chr3_-_100993507 | 0.15 |
ENST00000284322.10
|
ABI3BP
|
ABI family member 3 binding protein |
| chr14_-_20305932 | 0.15 |
ENST00000258821.8
ENST00000553828.1 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr3_-_100993409 | 0.15 |
ENST00000471714.6
|
ABI3BP
|
ABI family member 3 binding protein |
| chr17_-_41382298 | 0.15 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
| chr4_-_89057156 | 0.15 |
ENST00000511976.5
ENST00000509094.5 ENST00000264344.10 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13 member A |
| chr5_-_22853320 | 0.15 |
ENST00000504376.6
ENST00000382254.6 |
CDH12
|
cadherin 12 |
| chr3_+_4493442 | 0.14 |
ENST00000456211.8
ENST00000443694.5 ENST00000648266.1 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr2_+_188974364 | 0.14 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr18_-_55588535 | 0.14 |
ENST00000566286.5
ENST00000566279.5 ENST00000626595.2 ENST00000564999.5 ENST00000616053.4 ENST00000356073.8 |
TCF4
|
transcription factor 4 |
| chr7_-_64563063 | 0.14 |
ENST00000309683.11
|
ZNF680
|
zinc finger protein 680 |
| chrX_-_63785149 | 0.14 |
ENST00000671741.2
ENST00000625116.3 ENST00000624355.1 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr3_-_157160751 | 0.14 |
ENST00000461804.5
|
CCNL1
|
cyclin L1 |
| chr3_-_15797930 | 0.13 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr7_-_50565381 | 0.13 |
ENST00000444124.7
|
DDC
|
dopa decarboxylase |
| chr4_-_109801978 | 0.13 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr4_+_48483324 | 0.13 |
ENST00000273861.5
|
SLC10A4
|
solute carrier family 10 member 4 |
| chr3_+_185363129 | 0.13 |
ENST00000265026.8
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr6_+_125956696 | 0.13 |
ENST00000229633.7
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr3_-_71360753 | 0.13 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1 |
| chrX_+_119072960 | 0.13 |
ENST00000620151.3
|
ENSG00000277535.3
|
novel cancer/testis antigen family protein |
| chr3_-_123620571 | 0.13 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr3_-_123620496 | 0.12 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr17_+_11598456 | 0.12 |
ENST00000579828.5
ENST00000262442.9 ENST00000454412.6 |
DNAH9
|
dynein axonemal heavy chain 9 |
| chr1_+_43300971 | 0.12 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr10_-_50251500 | 0.12 |
ENST00000682911.1
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chrX_+_9463272 | 0.12 |
ENST00000407597.7
ENST00000380961.5 ENST00000424279.6 |
TBL1X
|
transducin beta like 1 X-linked |
| chr3_-_11568764 | 0.11 |
ENST00000424529.6
|
VGLL4
|
vestigial like family member 4 |
| chr21_-_28992947 | 0.11 |
ENST00000389194.7
ENST00000389195.7 ENST00000614971.4 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr3_-_146528750 | 0.11 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr2_-_151971750 | 0.11 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr1_+_200027605 | 0.11 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr3_-_15798184 | 0.11 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr2_+_186694007 | 0.11 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr10_-_20897288 | 0.11 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr7_+_150368189 | 0.11 |
ENST00000519397.1
ENST00000479668.5 |
REPIN1
|
replication initiator 1 |
| chr1_+_47023659 | 0.11 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1 |
| chr7_+_23710326 | 0.11 |
ENST00000354639.7
ENST00000531170.5 ENST00000444333.2 |
STK31
|
serine/threonine kinase 31 |
| chr1_+_100352451 | 0.11 |
ENST00000361544.11
ENST00000370124.8 ENST00000336454.5 |
CDC14A
|
cell division cycle 14A |
| chrX_-_117973579 | 0.11 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr4_+_94757921 | 0.10 |
ENST00000515059.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr12_+_45216079 | 0.10 |
ENST00000423947.7
ENST00000680498.1 ENST00000320560.13 |
ANO6
|
anoctamin 6 |
| chr7_+_23710203 | 0.10 |
ENST00000422637.5
ENST00000355870.8 |
STK31
|
serine/threonine kinase 31 |
| chr21_-_28992815 | 0.10 |
ENST00000361371.10
|
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr3_+_4680617 | 0.10 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr1_+_40396766 | 0.10 |
ENST00000539317.2
|
SMAP2
|
small ArfGAP2 |
| chr5_+_69565122 | 0.10 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr9_-_86947496 | 0.10 |
ENST00000298743.9
|
GAS1
|
growth arrest specific 1 |
| chr4_-_1208825 | 0.10 |
ENST00000511679.5
ENST00000617421.4 |
SPON2
|
spondin 2 |
| chr20_+_56629296 | 0.10 |
ENST00000201031.3
|
TFAP2C
|
transcription factor AP-2 gamma |
| chr3_+_57890011 | 0.10 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chrX_+_22136552 | 0.09 |
ENST00000682888.1
ENST00000684356.1 |
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr1_-_221742074 | 0.09 |
ENST00000366899.4
|
DUSP10
|
dual specificity phosphatase 10 |
| chr12_+_45216128 | 0.09 |
ENST00000681156.1
|
ANO6
|
anoctamin 6 |
| chr16_+_84368492 | 0.09 |
ENST00000416219.6
ENST00000262429.9 |
ATP2C2
|
ATPase secretory pathway Ca2+ transporting 2 |
| chr3_+_190615308 | 0.09 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr4_-_147684114 | 0.09 |
ENST00000322396.7
|
PRMT9
|
protein arginine methyltransferase 9 |
| chr1_+_15236509 | 0.09 |
ENST00000683790.1
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr8_-_9150648 | 0.09 |
ENST00000310455.4
|
PPP1R3B
|
protein phosphatase 1 regulatory subunit 3B |
| chr7_-_75994574 | 0.09 |
ENST00000439537.5
ENST00000493111.7 ENST00000417509.5 ENST00000485200.1 |
TMEM120A
|
transmembrane protein 120A |
| chr5_+_68288346 | 0.09 |
ENST00000320694.12
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr3_-_195583931 | 0.09 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr17_-_41397600 | 0.09 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr11_-_4697831 | 0.09 |
ENST00000641159.1
ENST00000396950.4 ENST00000532598.1 |
OR51C1P
OR51E2
|
olfactory receptor family 51 subfamily C member 1 pseudogene olfactory receptor family 51 subfamily E member 2 |
| chrX_+_118346072 | 0.09 |
ENST00000371822.9
ENST00000254029.8 ENST00000371825.7 |
WDR44
|
WD repeat domain 44 |
| chr3_-_157160094 | 0.09 |
ENST00000295925.5
ENST00000295926.8 |
CCNL1
|
cyclin L1 |
| chr3_-_126056786 | 0.09 |
ENST00000383598.6
|
SLC41A3
|
solute carrier family 41 member 3 |
| chr18_-_55588184 | 0.08 |
ENST00000354452.8
ENST00000565908.6 ENST00000635822.2 |
TCF4
|
transcription factor 4 |
| chr10_+_17951906 | 0.08 |
ENST00000377371.3
ENST00000377369.7 |
SLC39A12
|
solute carrier family 39 member 12 |
| chr3_+_132597260 | 0.08 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr17_+_57105899 | 0.08 |
ENST00000576295.5
|
AKAP1
|
A-kinase anchoring protein 1 |
| chr14_-_94323324 | 0.08 |
ENST00000341584.4
|
SERPINA6
|
serpin family A member 6 |
| chr12_+_69825273 | 0.08 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr10_+_17951885 | 0.08 |
ENST00000377374.8
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr1_-_207051202 | 0.08 |
ENST00000315927.9
|
YOD1
|
YOD1 deubiquitinase |
| chr3_+_98166696 | 0.08 |
ENST00000641450.1
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr3_-_132684685 | 0.08 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chr17_+_45160694 | 0.08 |
ENST00000307275.7
ENST00000585340.2 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chr4_+_20251896 | 0.07 |
ENST00000504154.6
|
SLIT2
|
slit guidance ligand 2 |
| chr19_-_48993300 | 0.07 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr6_-_52087569 | 0.07 |
ENST00000340994.4
ENST00000371117.8 |
PKHD1
|
PKHD1 ciliary IPT domain containing fibrocystin/polyductin |
| chr17_-_7329266 | 0.07 |
ENST00000571887.5
ENST00000315614.11 ENST00000399464.7 ENST00000570460.5 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr2_-_86105839 | 0.07 |
ENST00000263857.11
|
POLR1A
|
RNA polymerase I subunit A |
| chr20_+_21126037 | 0.07 |
ENST00000611685.4
ENST00000616848.4 |
KIZ
|
kizuna centrosomal protein |
| chr20_+_21125981 | 0.07 |
ENST00000619574.4
|
KIZ
|
kizuna centrosomal protein |
| chr1_-_111563956 | 0.07 |
ENST00000369717.8
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr6_+_133952162 | 0.07 |
ENST00000416965.5
ENST00000613034.4 |
TBPL1
|
TATA-box binding protein like 1 |
| chr17_-_37745018 | 0.07 |
ENST00000613727.4
ENST00000614313.4 ENST00000617811.5 ENST00000621123.4 |
HNF1B
|
HNF1 homeobox B |
| chr10_+_17951825 | 0.07 |
ENST00000539911.5
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr7_+_90383672 | 0.07 |
ENST00000416322.5
|
CLDN12
|
claudin 12 |
| chr10_-_112183698 | 0.07 |
ENST00000369425.5
ENST00000348367.9 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr13_-_36370171 | 0.07 |
ENST00000355182.8
|
SPART
|
spartin |
| chr12_-_39619782 | 0.07 |
ENST00000308666.4
|
ABCD2
|
ATP binding cassette subfamily D member 2 |
| chr19_+_35935550 | 0.07 |
ENST00000588831.5
|
LRFN3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr4_+_25655822 | 0.07 |
ENST00000504570.5
ENST00000382051.8 |
SLC34A2
|
solute carrier family 34 member 2 |
| chr20_+_21125999 | 0.07 |
ENST00000620891.4
|
KIZ
|
kizuna centrosomal protein |
| chr10_+_24449426 | 0.06 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217 |
| chr2_-_157327699 | 0.06 |
ENST00000397283.6
|
ERMN
|
ermin |
| chr8_-_92017292 | 0.06 |
ENST00000521553.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr13_-_36370111 | 0.06 |
ENST00000650221.1
|
SPART
|
spartin |
| chr2_+_169509693 | 0.06 |
ENST00000284669.2
|
KLHL41
|
kelch like family member 41 |
| chr8_-_94262308 | 0.06 |
ENST00000297596.3
ENST00000396194.6 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr3_-_18438767 | 0.06 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr11_-_33892010 | 0.06 |
ENST00000257818.3
|
LMO2
|
LIM domain only 2 |
| chrX_+_101623121 | 0.06 |
ENST00000491568.6
ENST00000479298.5 ENST00000471229.7 |
ARMCX3
|
armadillo repeat containing X-linked 3 |
| chr18_-_72865680 | 0.06 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr16_-_66730216 | 0.06 |
ENST00000569320.5
|
DYNC1LI2
|
dynein cytoplasmic 1 light intermediate chain 2 |
| chr17_-_58692032 | 0.06 |
ENST00000349033.10
ENST00000389934.7 |
TEX14
|
testis expressed 14, intercellular bridge forming factor |
| chr10_-_92243246 | 0.06 |
ENST00000412050.8
ENST00000614585.4 |
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr14_+_71586261 | 0.06 |
ENST00000358550.6
|
SIPA1L1
|
signal induced proliferation associated 1 like 1 |
| chr3_-_18424533 | 0.06 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr10_-_77638369 | 0.05 |
ENST00000372443.6
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr17_-_42018488 | 0.05 |
ENST00000589773.5
ENST00000674214.1 |
DNAJC7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr12_+_120694167 | 0.05 |
ENST00000535656.1
|
MLEC
|
malectin |
| chr7_-_73842508 | 0.05 |
ENST00000297873.9
|
METTL27
|
methyltransferase like 27 |
| chr20_-_25339731 | 0.05 |
ENST00000450393.5
ENST00000491682.5 |
ABHD12
|
abhydrolase domain containing 12, lysophospholipase |
| chr5_+_102808057 | 0.05 |
ENST00000684043.1
ENST00000682407.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_-_46303589 | 0.05 |
ENST00000617190.5
|
LRRC41
|
leucine rich repeat containing 41 |
| chr1_+_50109788 | 0.05 |
ENST00000651258.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr14_+_60734727 | 0.05 |
ENST00000261245.9
ENST00000539616.6 |
MNAT1
|
MNAT1 component of CDK activating kinase |
| chr2_+_47783172 | 0.05 |
ENST00000540021.6
|
MSH6
|
mutS homolog 6 |
| chr17_+_7336502 | 0.05 |
ENST00000158762.8
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr8_-_27992663 | 0.05 |
ENST00000380385.6
ENST00000354914.8 |
SCARA5
|
scavenger receptor class A member 5 |
| chr12_+_119667859 | 0.05 |
ENST00000541640.5
|
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr1_-_31373067 | 0.05 |
ENST00000373713.7
|
FABP3
|
fatty acid binding protein 3 |
| chr6_+_30908178 | 0.05 |
ENST00000259895.9
ENST00000376316.5 |
GTF2H4
|
general transcription factor IIH subunit 4 |
| chr19_+_859654 | 0.05 |
ENST00000592860.2
ENST00000327726.11 |
CFD
|
complement factor D |
| chr5_-_90409720 | 0.05 |
ENST00000522864.5
ENST00000283122.8 ENST00000522083.5 ENST00000522565.5 ENST00000522842.1 |
CETN3
|
centrin 3 |
| chr15_-_52652031 | 0.05 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr16_-_11782552 | 0.05 |
ENST00000396516.6
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr12_+_69825221 | 0.05 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr15_-_76012390 | 0.05 |
ENST00000394907.8
|
NRG4
|
neuregulin 4 |
| chr19_-_11346486 | 0.05 |
ENST00000590482.5
|
TMEM205
|
transmembrane protein 205 |
| chr14_-_49852760 | 0.05 |
ENST00000555970.5
ENST00000298310.10 ENST00000554626.5 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr1_+_78045956 | 0.04 |
ENST00000370759.4
|
GIPC2
|
GIPC PDZ domain containing family member 2 |
| chr1_+_50106265 | 0.04 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr17_-_58692021 | 0.04 |
ENST00000240361.12
|
TEX14
|
testis expressed 14, intercellular bridge forming factor |
| chr1_+_39215255 | 0.04 |
ENST00000671089.1
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr10_-_125775821 | 0.04 |
ENST00000368808.3
|
MMP21
|
matrix metallopeptidase 21 |
| chr16_-_18896874 | 0.04 |
ENST00000565324.5
ENST00000561947.5 |
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr20_+_45792498 | 0.04 |
ENST00000415790.5
|
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1 |
| chr11_+_122838492 | 0.04 |
ENST00000227348.9
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr11_+_34438900 | 0.04 |
ENST00000241052.5
|
CAT
|
catalase |
| chr5_+_36608146 | 0.04 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.5 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.4 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 1.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.3 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0001534 | radial spoke(GO:0001534) axonemal outer doublet(GO:0097545) |
| 0.1 | 0.5 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 1.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.3 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |