|
chr16_-_67393486
Show fit
|
3.17 |
ENST00000562206.1
ENST00000393957.7
ENST00000290942.9
|
TPPP3
|
tubulin polymerization promoting protein family member 3
|
|
chr2_-_153478753
Show fit
|
1.47 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog
|
|
chr15_-_93073111
Show fit
|
1.35 |
ENST00000557420.1
ENST00000542321.6
|
RGMA
|
repulsive guidance molecule BMP co-receptor a
|
|
chr13_+_42781547
Show fit
|
1.13 |
ENST00000537894.5
|
FAM216B
|
family with sequence similarity 216 member B
|
|
chr13_+_42781578
Show fit
|
1.12 |
ENST00000313851.3
|
FAM216B
|
family with sequence similarity 216 member B
|
|
chr1_-_66924791
Show fit
|
0.88 |
ENST00000371023.7
ENST00000371022.3
ENST00000371026.8
|
DNAI4
|
dynein axonemal intermediate chain 4
|
|
chr6_+_151239951
Show fit
|
0.86 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12
|
|
chr2_+_26401909
Show fit
|
0.74 |
ENST00000288710.7
|
DRC1
|
dynein regulatory complex subunit 1
|
|
chr10_-_77637558
Show fit
|
0.69 |
ENST00000372421.10
ENST00000639370.1
ENST00000640773.1
ENST00000638895.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr9_+_137241277
Show fit
|
0.57 |
ENST00000340384.5
|
TUBB4B
|
tubulin beta 4B class IVb
|
|
chr10_-_77637902
Show fit
|
0.57 |
ENST00000286627.10
ENST00000639486.1
ENST00000640523.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chrX_+_96684801
Show fit
|
0.50 |
ENST00000324765.13
|
DIAPH2
|
diaphanous related formin 2
|
|
chr22_-_30470577
Show fit
|
0.48 |
ENST00000435069.1
ENST00000540910.5
|
SEC14L3
|
SEC14 like lipid binding 3
|
|
chr10_-_77637444
Show fit
|
0.45 |
ENST00000639205.1
ENST00000639498.1
ENST00000372408.7
ENST00000372403.9
ENST00000640626.1
ENST00000404857.6
ENST00000638252.1
ENST00000640029.1
ENST00000640934.1
ENST00000639823.1
ENST00000372437.6
ENST00000639344.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr12_-_14885845
Show fit
|
0.42 |
ENST00000539261.6
ENST00000228938.5
|
MGP
|
matrix Gla protein
|
|
chrX_+_96684638
Show fit
|
0.36 |
ENST00000355827.8
ENST00000373061.7
|
DIAPH2
|
diaphanous related formin 2
|
|
chr6_+_122996227
Show fit
|
0.32 |
ENST00000275162.10
|
CLVS2
|
clavesin 2
|
|
chr4_-_89836213
Show fit
|
0.31 |
ENST00000618500.4
ENST00000508895.5
|
SNCA
|
synuclein alpha
|
|
chr13_+_35476740
Show fit
|
0.30 |
ENST00000537702.5
|
NBEA
|
neurobeachin
|
|
chrX_+_96684712
Show fit
|
0.29 |
ENST00000373049.8
|
DIAPH2
|
diaphanous related formin 2
|
|
chr1_-_223363337
Show fit
|
0.29 |
ENST00000608996.5
|
SUSD4
|
sushi domain containing 4
|
|
chr17_+_7855055
Show fit
|
0.28 |
ENST00000574668.1
ENST00000301599.7
|
TMEM88
|
transmembrane protein 88
|
|
chr12_+_57216779
Show fit
|
0.28 |
ENST00000349394.6
|
NXPH4
|
neurexophilin 4
|
|
chr9_+_26947039
Show fit
|
0.26 |
ENST00000519968.5
ENST00000433700.5
|
IFT74
|
intraflagellar transport 74
|
|
chr1_-_217076889
Show fit
|
0.26 |
ENST00000493748.5
ENST00000463665.5
|
ESRRG
|
estrogen related receptor gamma
|
|
chr8_+_103140692
Show fit
|
0.26 |
ENST00000438105.2
ENST00000309982.10
ENST00000297574.6
|
BAALC
|
BAALC binder of MAP3K1 and KLF4
|
|
chr10_-_77637789
Show fit
|
0.26 |
ENST00000481070.1
ENST00000640969.1
ENST00000286628.14
ENST00000638991.1
ENST00000639913.1
ENST00000480683.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr4_+_76435216
Show fit
|
0.26 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3
|
|
chr4_-_89835617
Show fit
|
0.25 |
ENST00000611107.1
ENST00000345009.8
ENST00000505199.5
ENST00000502987.5
|
SNCA
|
synuclein alpha
|
|
chr10_+_94089067
Show fit
|
0.23 |
ENST00000371375.1
ENST00000675218.1
|
PLCE1
|
phospholipase C epsilon 1
|
|
chr3_-_51779187
Show fit
|
0.22 |
ENST00000398780.5
ENST00000668964.1
ENST00000667863.2
ENST00000647442.1
|
IQCF6
|
IQ motif containing F6
|
|
chr10_+_94089034
Show fit
|
0.22 |
ENST00000676102.1
ENST00000371385.8
|
PLCE1
|
phospholipase C epsilon 1
|
|
chr12_-_57078739
Show fit
|
0.22 |
ENST00000379391.7
|
NEMP1
|
nuclear envelope integral membrane protein 1
|
|
chr8_-_142879820
Show fit
|
0.22 |
ENST00000377675.3
ENST00000517471.5
ENST00000292427.10
|
CYP11B1
|
cytochrome P450 family 11 subfamily B member 1
|
|
chrX_-_10677720
Show fit
|
0.22 |
ENST00000453318.6
|
MID1
|
midline 1
|
|
chr2_-_212124901
Show fit
|
0.22 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
chr14_-_21048431
Show fit
|
0.22 |
ENST00000555026.5
|
NDRG2
|
NDRG family member 2
|
|
chr7_-_144259722
Show fit
|
0.21 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor family 2 subfamily A member 7
|
|
chr10_-_77637721
Show fit
|
0.20 |
ENST00000638848.1
ENST00000639406.1
ENST00000618048.2
ENST00000639120.1
ENST00000640834.1
ENST00000639601.1
ENST00000638514.1
ENST00000457953.6
ENST00000639090.1
ENST00000639489.1
ENST00000372440.6
ENST00000404771.8
ENST00000638203.1
ENST00000638306.1
ENST00000638351.1
ENST00000638606.1
ENST00000639591.1
ENST00000640182.1
ENST00000640605.1
ENST00000640141.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr11_+_31816266
Show fit
|
0.20 |
ENST00000644607.1
ENST00000646221.1
ENST00000643671.1
ENST00000643931.1
ENST00000642614.1
ENST00000642818.1
ENST00000645848.1
ENST00000506388.2
ENST00000645824.1
ENST00000532942.5
|
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA
novel protein
|
|
chr12_-_57078784
Show fit
|
0.20 |
ENST00000300128.9
|
NEMP1
|
nuclear envelope integral membrane protein 1
|
|
chr18_+_59220149
Show fit
|
0.19 |
ENST00000256857.7
ENST00000529320.2
ENST00000420468.6
|
GRP
|
gastrin releasing peptide
|
|
chr1_-_91886144
Show fit
|
0.19 |
ENST00000212355.9
|
TGFBR3
|
transforming growth factor beta receptor 3
|
|
chr1_-_182672232
Show fit
|
0.19 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8
|
|
chr11_-_111911759
Show fit
|
0.19 |
ENST00000650687.2
|
CRYAB
|
crystallin alpha B
|
|
chr1_-_107965009
Show fit
|
0.18 |
ENST00000527011.5
ENST00000370056.9
|
VAV3
|
vav guanine nucleotide exchange factor 3
|
|
chrX_+_126819720
Show fit
|
0.18 |
ENST00000371125.4
|
PRR32
|
proline rich 32
|
|
chr16_+_15502266
Show fit
|
0.17 |
ENST00000452191.6
|
BMERB1
|
bMERB domain containing 1
|
|
chr1_-_182672882
Show fit
|
0.16 |
ENST00000367557.8
ENST00000258302.8
|
RGS8
|
regulator of G protein signaling 8
|
|
chr7_-_27130733
Show fit
|
0.16 |
ENST00000428284.2
ENST00000360046.10
ENST00000610970.1
|
HOXA4
|
homeobox A4
|
|
chr17_-_1684807
Show fit
|
0.16 |
ENST00000577001.1
ENST00000572621.5
ENST00000304992.11
|
PRPF8
|
pre-mRNA processing factor 8
|
|
chr17_-_44199206
Show fit
|
0.15 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7 like 3
|
|
chr7_-_144410227
Show fit
|
0.15 |
ENST00000467773.1
ENST00000483238.5
|
NOBOX
|
NOBOX oogenesis homeobox
|
|
chr13_-_77919459
Show fit
|
0.15 |
ENST00000643890.1
|
EDNRB
|
endothelin receptor type B
|
|
chr3_+_156142962
Show fit
|
0.15 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1
|
|
chr1_-_43367689
Show fit
|
0.14 |
ENST00000621943.4
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr10_-_97185920
Show fit
|
0.14 |
ENST00000371041.3
ENST00000266058.9
|
SLIT1
|
slit guidance ligand 1
|
|
chr5_+_167754918
Show fit
|
0.14 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2
|
|
chr12_+_56021005
Show fit
|
0.14 |
ENST00000547167.6
|
IKZF4
|
IKAROS family zinc finger 4
|
|
chr10_+_24466487
Show fit
|
0.14 |
ENST00000396446.5
ENST00000396445.5
ENST00000376451.4
|
KIAA1217
|
KIAA1217
|
|
chr6_+_68635273
Show fit
|
0.13 |
ENST00000370598.6
|
ADGRB3
|
adhesion G protein-coupled receptor B3
|
|
chr7_-_78771265
Show fit
|
0.13 |
ENST00000630991.2
ENST00000629359.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr9_-_124507382
Show fit
|
0.13 |
ENST00000373588.9
ENST00000620110.4
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1
|
|
chr1_-_43368039
Show fit
|
0.13 |
ENST00000413844.3
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr5_+_163437569
Show fit
|
0.12 |
ENST00000512163.5
ENST00000393929.5
ENST00000510097.5
ENST00000340828.7
ENST00000511490.4
ENST00000510664.5
|
CCNG1
|
cyclin G1
|
|
chr1_-_43367956
Show fit
|
0.12 |
ENST00000372458.8
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr6_+_33420052
Show fit
|
0.12 |
ENST00000646630.1
ENST00000629380.3
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr7_-_78771108
Show fit
|
0.12 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr3_-_150546403
Show fit
|
0.11 |
ENST00000239944.7
ENST00000491660.1
ENST00000487153.1
|
SERP1
|
stress associated endoplasmic reticulum protein 1
|
|
chr10_-_77637633
Show fit
|
0.11 |
ENST00000638223.1
ENST00000639544.1
ENST00000640807.1
ENST00000434208.6
ENST00000626620.3
ENST00000638575.1
ENST00000638759.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr3_+_148827800
Show fit
|
0.11 |
ENST00000282957.9
ENST00000468341.1
|
CPB1
|
carboxypeptidase B1
|
|
chr17_+_60677822
Show fit
|
0.11 |
ENST00000407086.8
ENST00000589222.5
ENST00000626960.2
ENST00000390652.9
|
BCAS3
|
BCAS3 microtubule associated cell migration factor
|
|
chr6_+_33420201
Show fit
|
0.10 |
ENST00000644458.1
ENST00000449372.7
ENST00000628646.2
ENST00000418600.7
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chrX_+_77447387
Show fit
|
0.09 |
ENST00000439435.3
|
FGF16
|
fibroblast growth factor 16
|
|
chr16_-_49856105
Show fit
|
0.09 |
ENST00000563137.7
|
ZNF423
|
zinc finger protein 423
|
|
chr4_-_5019437
Show fit
|
0.09 |
ENST00000506508.1
ENST00000509419.1
ENST00000307746.9
|
CYTL1
|
cytokine like 1
|
|
chr17_+_45161864
Show fit
|
0.09 |
ENST00000589230.6
ENST00000591576.5
ENST00000591070.6
ENST00000592695.1
|
HEXIM2
|
HEXIM P-TEFb complex subunit 2
|
|
chr2_-_49154433
Show fit
|
0.08 |
ENST00000454032.5
ENST00000304421.8
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr10_+_18400562
Show fit
|
0.08 |
ENST00000377315.5
ENST00000650685.1
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr7_-_126533850
Show fit
|
0.08 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8
|
|
chr16_+_30069539
Show fit
|
0.08 |
ENST00000565355.1
|
ALDOA
|
aldolase, fructose-bisphosphate A
|
|
chr15_+_52019206
Show fit
|
0.08 |
ENST00000681888.1
ENST00000261845.7
ENST00000680066.1
ENST00000680777.1
ENST00000680652.1
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr2_-_49154507
Show fit
|
0.08 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr6_-_131701401
Show fit
|
0.08 |
ENST00000315453.4
|
OR2A4
|
olfactory receptor family 2 subfamily A member 4
|
|
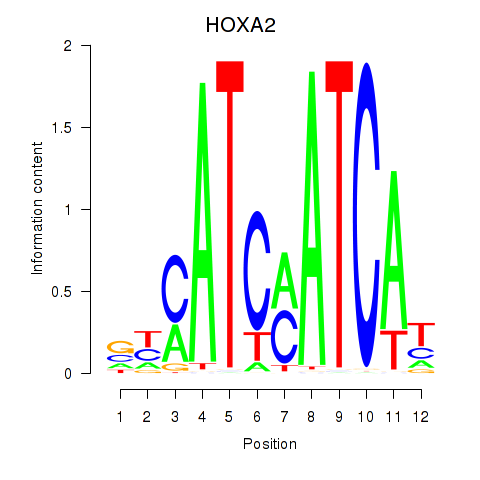
chr7_-_27102669
Show fit
|
0.07 |
ENST00000222718.7
|
HOXA2
|
homeobox A2
|
|
chr14_+_22516273
Show fit
|
0.07 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27
|
|
chr8_+_142449430
Show fit
|
0.07 |
ENST00000643448.1
ENST00000517894.5
|
ADGRB1
|
adhesion G protein-coupled receptor B1
|
|
chr17_-_48545077
Show fit
|
0.07 |
ENST00000330070.6
|
HOXB2
|
homeobox B2
|
|
chr7_-_101237827
Show fit
|
0.07 |
ENST00000611078.4
|
CLDN15
|
claudin 15
|
|
chr12_+_56007484
Show fit
|
0.06 |
ENST00000262032.9
|
IKZF4
|
IKAROS family zinc finger 4
|
|
chr15_+_47717344
Show fit
|
0.06 |
ENST00000558816.5
ENST00000536845.7
|
SEMA6D
|
semaphorin 6D
|
|
chr2_-_152098810
Show fit
|
0.06 |
ENST00000636442.1
ENST00000638005.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4
|
|
chr17_-_39197696
Show fit
|
0.06 |
ENST00000394310.7
ENST00000622445.4
ENST00000344140.5
|
CACNB1
|
calcium voltage-gated channel auxiliary subunit beta 1
|
|
chr7_+_33904911
Show fit
|
0.05 |
ENST00000297161.6
|
BMPER
|
BMP binding endothelial regulator
|
|
chr5_-_179617581
Show fit
|
0.05 |
ENST00000523449.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr15_-_64381431
Show fit
|
0.05 |
ENST00000558008.3
ENST00000300035.9
ENST00000559519.5
ENST00000380258.6
|
PCLAF
|
PCNA clamp associated factor
|
|
chr14_-_81427390
Show fit
|
0.05 |
ENST00000555447.5
|
STON2
|
stonin 2
|
|
chr11_-_2140967
Show fit
|
0.05 |
ENST00000381389.5
|
IGF2
|
insulin like growth factor 2
|
|
chr7_-_13989658
Show fit
|
0.04 |
ENST00000430479.6
ENST00000433547.1
ENST00000405192.6
|
ETV1
|
ETS variant transcription factor 1
|
|
chr11_-_123058991
Show fit
|
0.04 |
ENST00000526686.1
|
HSPA8
|
heat shock protein family A (Hsp70) member 8
|
|
chr18_-_55403682
Show fit
|
0.04 |
ENST00000564228.5
ENST00000630828.2
|
TCF4
|
transcription factor 4
|
|
chr6_-_32128191
Show fit
|
0.04 |
ENST00000453203.2
ENST00000375203.8
ENST00000375201.8
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr1_-_160579439
Show fit
|
0.04 |
ENST00000368054.8
ENST00000368048.7
ENST00000311224.8
ENST00000368051.3
ENST00000534968.5
|
CD84
|
CD84 molecule
|
|
chr11_-_123059413
Show fit
|
0.03 |
ENST00000524552.5
|
HSPA8
|
heat shock protein family A (Hsp70) member 8
|
|
chr7_-_120857124
Show fit
|
0.03 |
ENST00000441017.5
ENST00000424710.5
ENST00000433758.5
|
TSPAN12
|
tetraspanin 12
|
|
chr10_-_63269057
Show fit
|
0.03 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr8_-_78805306
Show fit
|
0.03 |
ENST00000639719.1
ENST00000263851.9
|
IL7
|
interleukin 7
|
|
chr9_+_110668854
Show fit
|
0.03 |
ENST00000189978.10
ENST00000374440.7
|
MUSK
|
muscle associated receptor tyrosine kinase
|
|
chr7_-_13989891
Show fit
|
0.03 |
ENST00000405218.6
|
ETV1
|
ETS variant transcription factor 1
|
|
chr1_+_243256034
Show fit
|
0.03 |
ENST00000366541.8
|
SDCCAG8
|
SHH signaling and ciliogenesis regulator SDCCAG8
|
|
chr10_+_35175586
Show fit
|
0.03 |
ENST00000494479.5
ENST00000463314.5
ENST00000342105.7
ENST00000495301.1
|
CREM
|
cAMP responsive element modulator
|
|
chr2_-_144520315
Show fit
|
0.03 |
ENST00000465070.5
ENST00000465308.5
ENST00000636471.1
ENST00000629520.2
ENST00000675069.1
ENST00000636026.2
ENST00000444559.5
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr4_-_108168919
Show fit
|
0.03 |
ENST00000265165.6
|
LEF1
|
lymphoid enhancer binding factor 1
|
|
chr17_-_39197652
Show fit
|
0.03 |
ENST00000394303.8
|
CACNB1
|
calcium voltage-gated channel auxiliary subunit beta 1
|
|
chr9_+_110668779
Show fit
|
0.03 |
ENST00000416899.7
ENST00000374448.9
|
MUSK
|
muscle associated receptor tyrosine kinase
|
|
chr17_+_81528370
Show fit
|
0.02 |
ENST00000417245.7
ENST00000334850.7
|
FSCN2
|
fascin actin-bundling protein 2, retinal
|
|
chr1_+_64470120
Show fit
|
0.02 |
ENST00000651257.2
|
CACHD1
|
cache domain containing 1
|
|
chr8_-_78805515
Show fit
|
0.02 |
ENST00000379113.6
ENST00000541183.2
|
IL7
|
interleukin 7
|
|
chr11_-_127000762
Show fit
|
0.02 |
ENST00000525144.7
|
KIRREL3
|
kirre like nephrin family adhesion molecule 3
|
|
chr12_-_88029385
Show fit
|
0.02 |
ENST00000298699.7
ENST00000550553.5
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr11_+_26188836
Show fit
|
0.02 |
ENST00000672621.1
|
ANO3
|
anoctamin 3
|
|
chr19_+_35358821
Show fit
|
0.02 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3
|
|
chr4_-_108168950
Show fit
|
0.02 |
ENST00000379951.6
|
LEF1
|
lymphoid enhancer binding factor 1
|
|
chrX_+_100644183
Show fit
|
0.02 |
ENST00000640889.1
ENST00000373004.5
|
SRPX2
|
sushi repeat containing protein X-linked 2
|
|
chr12_+_57089094
Show fit
|
0.01 |
ENST00000342556.6
ENST00000300131.8
|
NAB2
|
NGFI-A binding protein 2
|
|
chrX_-_111412162
Show fit
|
0.01 |
ENST00000637570.1
ENST00000356220.8
ENST00000636035.2
ENST00000637453.1
ENST00000635795.1
|
DCX
|
doublecortin
|
|
chr12_-_48999363
Show fit
|
0.01 |
ENST00000421952.3
|
DDN
|
dendrin
|
|
chr7_+_20330893
Show fit
|
0.01 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8
|
|
chr20_+_36154630
Show fit
|
0.01 |
ENST00000338074.7
ENST00000636016.2
ENST00000373945.5
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chrX_+_48597482
Show fit
|
0.01 |
ENST00000218056.9
ENST00000376729.10
|
WDR13
|
WD repeat domain 13
|
|
chr3_+_189789672
Show fit
|
0.01 |
ENST00000434928.5
|
TP63
|
tumor protein p63
|
|
chr1_-_109041986
Show fit
|
0.01 |
ENST00000400794.7
ENST00000528747.1
ENST00000361054.7
|
WDR47
|
WD repeat domain 47
|
|
chr21_+_18244828
Show fit
|
0.00 |
ENST00000299295.7
|
CHODL
|
chondrolectin
|
|
chr20_+_36214373
Show fit
|
0.00 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chrX_-_66033664
Show fit
|
0.00 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4
|
|
chr17_+_39668447
Show fit
|
0.00 |
ENST00000269582.3
ENST00000581428.1
|
PNMT
|
phenylethanolamine N-methyltransferase
|
|
chr8_+_94641074
Show fit
|
0.00 |
ENST00000423620.6
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr21_-_42010327
Show fit
|
0.00 |
ENST00000398505.7
ENST00000449949.5
ENST00000310826.10
ENST00000398499.5
ENST00000398497.2
ENST00000398511.3
|
ZBTB21
|
zinc finger and BTB domain containing 21
|
|
chrX_+_70062457
Show fit
|
0.00 |
ENST00000338352.3
|
OTUD6A
|
OTU deubiquitinase 6A
|