Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
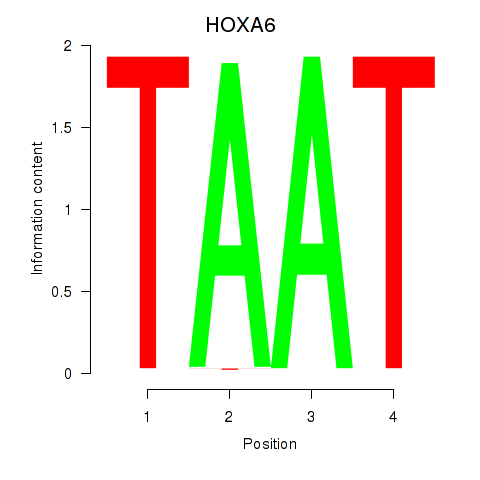
Results for HOXA6
Z-value: 0.51
Transcription factors associated with HOXA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA6
|
ENSG00000106006.6 | HOXA6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA6 | hg38_v1_chr7_-_27147774_27147774 | -0.03 | 8.8e-01 | Click! |
Activity profile of HOXA6 motif
Sorted Z-values of HOXA6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_27143672 | 0.96 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr3_+_41194741 | 0.89 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr1_-_152159227 | 0.88 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr14_-_53956811 | 0.75 |
ENST00000559087.5
ENST00000245451.9 |
BMP4
|
bone morphogenetic protein 4 |
| chr12_-_91182652 | 0.72 |
ENST00000552145.5
ENST00000546745.5 |
DCN
|
decorin |
| chr13_-_94479671 | 0.61 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr13_+_35476740 | 0.59 |
ENST00000537702.5
|
NBEA
|
neurobeachin |
| chr9_+_2159850 | 0.57 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_61404076 | 0.55 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr9_+_2159672 | 0.54 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_124746630 | 0.51 |
ENST00000513986.2
|
ZNF608
|
zinc finger protein 608 |
| chr5_-_88883701 | 0.48 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr13_-_85799400 | 0.47 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr19_+_13023958 | 0.47 |
ENST00000587760.5
ENST00000585575.5 |
NFIX
|
nuclear factor I X |
| chr17_-_40755328 | 0.45 |
ENST00000312150.5
|
KRT25
|
keratin 25 |
| chr18_-_55422306 | 0.43 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr6_+_104957099 | 0.43 |
ENST00000345080.5
|
LIN28B
|
lin-28 homolog B |
| chr17_-_40937445 | 0.40 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr12_-_91182784 | 0.39 |
ENST00000547568.6
ENST00000052754.10 ENST00000552962.5 |
DCN
|
decorin |
| chr9_+_2717479 | 0.38 |
ENST00000382082.4
|
KCNV2
|
potassium voltage-gated channel modifier subfamily V member 2 |
| chr5_+_36606355 | 0.37 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr9_+_2015275 | 0.36 |
ENST00000637103.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_-_54928044 | 0.36 |
ENST00000273286.6
|
LRTM1
|
leucine rich repeats and transmembrane domains 1 |
| chr9_+_2015186 | 0.35 |
ENST00000357248.8
ENST00000450198.6 ENST00000634287.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_13095664 | 0.35 |
ENST00000337630.10
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr4_+_159267737 | 0.34 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr8_+_76683779 | 0.33 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr12_+_41437680 | 0.33 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr18_-_55422492 | 0.33 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr15_-_48963912 | 0.33 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4 |
| chr7_-_78489900 | 0.32 |
ENST00000636039.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_+_2015335 | 0.31 |
ENST00000636559.1
ENST00000349721.8 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_-_40937641 | 0.31 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr1_+_60865259 | 0.30 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr4_-_173530219 | 0.29 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr5_-_1882902 | 0.29 |
ENST00000231357.7
|
IRX4
|
iroquois homeobox 4 |
| chr5_-_124745315 | 0.29 |
ENST00000306315.9
|
ZNF608
|
zinc finger protein 608 |
| chr17_-_55511434 | 0.28 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36 |
| chr14_-_91947383 | 0.27 |
ENST00000267620.14
|
FBLN5
|
fibulin 5 |
| chr1_+_61082702 | 0.27 |
ENST00000485903.6
ENST00000371185.6 ENST00000371184.6 |
NFIA
|
nuclear factor I A |
| chr10_+_123135938 | 0.27 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr17_-_48593748 | 0.26 |
ENST00000239151.6
|
HOXB5
|
homeobox B5 |
| chr5_-_88785493 | 0.26 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr5_-_88883420 | 0.24 |
ENST00000437473.6
|
MEF2C
|
myocyte enhancer factor 2C |
| chr16_+_53099100 | 0.23 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr3_-_114758940 | 0.23 |
ENST00000464560.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_61082553 | 0.23 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr11_-_33892010 | 0.23 |
ENST00000257818.3
|
LMO2
|
LIM domain only 2 |
| chr16_+_53207981 | 0.22 |
ENST00000565803.2
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr7_-_5425404 | 0.22 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr14_+_22508602 | 0.22 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr11_+_123525822 | 0.22 |
ENST00000322282.11
ENST00000529750.5 |
GRAMD1B
|
GRAM domain containing 1B |
| chr7_+_114414809 | 0.21 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr7_-_27165517 | 0.21 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr9_-_14722725 | 0.21 |
ENST00000380911.4
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr7_-_126533850 | 0.20 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr9_+_2158487 | 0.20 |
ENST00000634706.1
ENST00000634338.1 ENST00000635688.1 ENST00000634435.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_129375643 | 0.19 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr21_-_34526850 | 0.19 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr3_-_114759115 | 0.19 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr14_-_36582593 | 0.19 |
ENST00000258829.6
|
NKX2-8
|
NK2 homeobox 8 |
| chrX_-_124963768 | 0.19 |
ENST00000371130.7
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr8_+_103819244 | 0.19 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_87331668 | 0.18 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr2_-_80304274 | 0.18 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr13_-_45851730 | 0.18 |
ENST00000400405.4
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr18_-_26657401 | 0.18 |
ENST00000580191.5
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr3_+_141386862 | 0.18 |
ENST00000513258.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_-_76072654 | 0.18 |
ENST00000369950.8
ENST00000611179.4 ENST00000369963.5 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr7_-_13986439 | 0.18 |
ENST00000443608.5
ENST00000438956.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr3_-_114624193 | 0.17 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr15_-_77696142 | 0.17 |
ENST00000561030.5
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr3_-_65597886 | 0.17 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr5_-_77638713 | 0.16 |
ENST00000306422.5
|
OTP
|
orthopedia homeobox |
| chr1_+_40709475 | 0.16 |
ENST00000372651.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr16_+_53208438 | 0.16 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr10_-_77090722 | 0.16 |
ENST00000638531.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr10_+_47322450 | 0.16 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr12_-_23951020 | 0.16 |
ENST00000441133.2
ENST00000545921.5 |
SOX5
|
SRY-box transcription factor 5 |
| chr4_-_115113822 | 0.16 |
ENST00000613194.4
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr2_-_213151590 | 0.16 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr1_-_203086001 | 0.16 |
ENST00000241651.5
|
MYOG
|
myogenin |
| chr16_-_51151259 | 0.15 |
ENST00000251020.9
|
SALL1
|
spalt like transcription factor 1 |
| chr4_+_41612892 | 0.15 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr8_+_103880412 | 0.15 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr5_+_173889337 | 0.15 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_-_152352800 | 0.15 |
ENST00000393956.9
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr5_+_174724549 | 0.15 |
ENST00000239243.7
ENST00000507785.2 |
MSX2
|
msh homeobox 2 |
| chr4_+_41612702 | 0.14 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_+_114414997 | 0.14 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr15_-_53733103 | 0.14 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr18_+_61333424 | 0.14 |
ENST00000262717.9
|
CDH20
|
cadherin 20 |
| chr5_-_60488055 | 0.14 |
ENST00000505507.6
ENST00000515835.2 ENST00000502484.6 |
PDE4D
|
phosphodiesterase 4D |
| chr17_-_10518536 | 0.14 |
ENST00000226207.6
|
MYH1
|
myosin heavy chain 1 |
| chr15_+_58138368 | 0.14 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr4_-_109302643 | 0.13 |
ENST00000399126.1
ENST00000505591.1 ENST00000399132.6 |
COL25A1
|
collagen type XXV alpha 1 chain |
| chr8_+_84705920 | 0.13 |
ENST00000523850.5
ENST00000521376.1 |
RALYL
|
RALY RNA binding protein like |
| chr4_-_109302707 | 0.13 |
ENST00000642955.1
|
COL25A1
|
collagen type XXV alpha 1 chain |
| chr6_-_55875583 | 0.13 |
ENST00000370830.4
|
BMP5
|
bone morphogenetic protein 5 |
| chr20_+_33562306 | 0.13 |
ENST00000344201.7
|
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr5_-_88883147 | 0.13 |
ENST00000513252.5
ENST00000506554.5 ENST00000508569.5 ENST00000637732.1 ENST00000504921.7 ENST00000637481.1 ENST00000510942.5 |
MEF2C
|
myocyte enhancer factor 2C |
| chr7_+_114416286 | 0.13 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2 |
| chrX_-_32412220 | 0.13 |
ENST00000619831.5
|
DMD
|
dystrophin |
| chr16_+_55655960 | 0.13 |
ENST00000568943.6
|
SLC6A2
|
solute carrier family 6 member 2 |
| chr5_+_177384430 | 0.13 |
ENST00000512593.5
ENST00000324417.6 |
SLC34A1
|
solute carrier family 34 member 1 |
| chr5_+_139308095 | 0.13 |
ENST00000515833.2
|
MATR3
|
matrin 3 |
| chr7_-_5423543 | 0.13 |
ENST00000399537.8
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr9_-_121050264 | 0.13 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr4_-_175812746 | 0.13 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A |
| chr2_+_170178136 | 0.13 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr2_-_29074515 | 0.13 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr4_-_115113614 | 0.12 |
ENST00000264363.7
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr2_-_99255107 | 0.12 |
ENST00000333017.6
ENST00000626374.2 ENST00000409679.5 ENST00000423306.1 |
LYG2
|
lysozyme g2 |
| chr16_+_55656249 | 0.12 |
ENST00000219833.13
ENST00000574918.2 |
SLC6A2
|
solute carrier family 6 member 2 |
| chr8_-_132111159 | 0.12 |
ENST00000673615.1
ENST00000434736.6 |
HHLA1
|
HERV-H LTR-associating 1 |
| chr1_+_50108856 | 0.12 |
ENST00000650764.1
ENST00000494555.2 ENST00000371824.7 ENST00000371823.8 ENST00000652693.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chrM_+_9207 | 0.12 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_-_48604959 | 0.12 |
ENST00000225648.4
ENST00000484302.3 |
HOXB6
|
homeobox B6 |
| chr9_-_70869076 | 0.12 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr20_+_64164446 | 0.12 |
ENST00000328439.6
|
MYT1
|
myelin transcription factor 1 |
| chrX_-_33128360 | 0.11 |
ENST00000378677.6
|
DMD
|
dystrophin |
| chr20_+_64164474 | 0.11 |
ENST00000622439.4
ENST00000536311.5 |
MYT1
|
myelin transcription factor 1 |
| chr2_-_45009401 | 0.11 |
ENST00000303077.7
|
SIX2
|
SIX homeobox 2 |
| chr17_-_48615261 | 0.11 |
ENST00000239144.5
|
HOXB8
|
homeobox B8 |
| chr9_-_20622479 | 0.11 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr4_+_128809791 | 0.11 |
ENST00000452328.6
ENST00000504089.5 |
JADE1
|
jade family PHD finger 1 |
| chr4_+_99574812 | 0.11 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr11_-_124320197 | 0.11 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr20_+_64164566 | 0.11 |
ENST00000650655.1
|
MYT1
|
myelin transcription factor 1 |
| chr18_-_32772879 | 0.11 |
ENST00000358095.4
ENST00000359358.9 |
KLHL14
|
kelch like family member 14 |
| chr5_+_161685708 | 0.11 |
ENST00000274545.10
|
GABRA6
|
gamma-aminobutyric acid type A receptor subunit alpha6 |
| chr9_+_2158239 | 0.11 |
ENST00000635133.1
ENST00000634931.1 ENST00000423555.6 ENST00000382185.6 ENST00000302401.8 ENST00000382183.6 ENST00000417599.6 ENST00000382186.6 ENST00000635530.1 ENST00000635388.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_34043205 | 0.11 |
ENST00000382065.8
ENST00000231338.7 |
C1QTNF3
|
C1q and TNF related 3 |
| chr9_-_70869029 | 0.11 |
ENST00000361823.9
ENST00000377101.5 ENST00000360823.6 ENST00000377105.5 |
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr5_+_161685748 | 0.11 |
ENST00000523217.5
|
GABRA6
|
gamma-aminobutyric acid type A receptor subunit alpha6 |
| chrM_+_5824 | 0.11 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr17_-_40994159 | 0.11 |
ENST00000391586.3
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr17_-_48545077 | 0.11 |
ENST00000330070.6
|
HOXB2
|
homeobox B2 |
| chr1_+_40709316 | 0.10 |
ENST00000372652.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr20_+_6007245 | 0.10 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr8_+_91249307 | 0.10 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr2_+_225399684 | 0.10 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr16_+_55656089 | 0.10 |
ENST00000414754.7
|
SLC6A2
|
solute carrier family 6 member 2 |
| chr20_-_51802433 | 0.10 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr2_+_66435558 | 0.10 |
ENST00000488550.5
|
MEIS1
|
Meis homeobox 1 |
| chr9_-_95509241 | 0.10 |
ENST00000331920.11
|
PTCH1
|
patched 1 |
| chr16_+_55656294 | 0.10 |
ENST00000379906.6
|
SLC6A2
|
solute carrier family 6 member 2 |
| chr20_+_33562365 | 0.10 |
ENST00000346541.7
ENST00000397800.5 ENST00000492345.5 |
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr2_-_181680490 | 0.10 |
ENST00000684145.1
ENST00000295108.4 ENST00000684079.1 ENST00000683430.1 |
CERKL
NEUROD1
|
ceramide kinase like neuronal differentiation 1 |
| chr11_-_16356538 | 0.10 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr7_-_27102669 | 0.10 |
ENST00000222718.7
|
HOXA2
|
homeobox A2 |
| chr12_-_99154492 | 0.10 |
ENST00000546568.5
ENST00000546960.5 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr8_-_115668966 | 0.10 |
ENST00000395715.8
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chrX_+_9463272 | 0.09 |
ENST00000407597.7
ENST00000380961.5 ENST00000424279.6 |
TBL1X
|
transducin beta like 1 X-linked |
| chr2_-_77522347 | 0.09 |
ENST00000409093.1
ENST00000409884.6 ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr2_-_2324323 | 0.09 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr12_-_91004965 | 0.09 |
ENST00000261172.8
|
EPYC
|
epiphycan |
| chr5_-_91383310 | 0.09 |
ENST00000265138.4
|
ARRDC3
|
arrestin domain containing 3 |
| chr9_+_79571956 | 0.09 |
ENST00000376552.8
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr15_+_96325935 | 0.09 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr5_-_27038576 | 0.09 |
ENST00000511822.1
ENST00000231021.9 |
CDH9
|
cadherin 9 |
| chr2_-_174846405 | 0.09 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr15_+_96333111 | 0.09 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr16_+_8712943 | 0.09 |
ENST00000561870.5
ENST00000396600.6 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr8_-_115668609 | 0.09 |
ENST00000220888.9
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr7_-_27140195 | 0.09 |
ENST00000522788.5
ENST00000317201.7 |
HOXA3
|
homeobox A3 |
| chr6_-_33192454 | 0.08 |
ENST00000395194.1
ENST00000341947.7 ENST00000374708.8 |
COL11A2
|
collagen type XI alpha 2 chain |
| chr5_-_38557459 | 0.08 |
ENST00000511561.1
|
LIFR
|
LIF receptor subunit alpha |
| chr3_-_27722699 | 0.08 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr13_+_48256214 | 0.08 |
ENST00000650237.1
|
ITM2B
|
integral membrane protein 2B |
| chr3_-_62374293 | 0.08 |
ENST00000486811.5
|
FEZF2
|
FEZ family zinc finger 2 |
| chr6_+_50713526 | 0.08 |
ENST00000008391.4
|
TFAP2D
|
transcription factor AP-2 delta |
| chr17_-_10549694 | 0.08 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chr5_-_161546708 | 0.08 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr3_+_69936583 | 0.08 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr2_+_170715317 | 0.08 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor |
| chr11_-_31811034 | 0.08 |
ENST00000638250.1
|
PAX6
|
paired box 6 |
| chr11_+_20599602 | 0.08 |
ENST00000525748.6
|
SLC6A5
|
solute carrier family 6 member 5 |
| chr16_+_2682515 | 0.07 |
ENST00000301738.9
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr3_+_69763726 | 0.07 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr7_-_5423826 | 0.07 |
ENST00000430969.6
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr3_-_62373538 | 0.07 |
ENST00000283268.8
|
FEZF2
|
FEZ family zinc finger 2 |
| chr6_-_84764581 | 0.07 |
ENST00000369663.10
|
TBX18
|
T-box transcription factor 18 |
| chr11_-_8263858 | 0.07 |
ENST00000534484.1
ENST00000335790.8 |
LMO1
|
LIM domain only 1 |
| chr2_-_182427014 | 0.07 |
ENST00000409365.5
ENST00000351439.9 |
PDE1A
|
phosphodiesterase 1A |
| chr17_+_76540035 | 0.07 |
ENST00000592014.6
|
PRCD
|
photoreceptor disc component |
| chr15_+_33968484 | 0.07 |
ENST00000383263.7
|
CHRM5
|
cholinergic receptor muscarinic 5 |
| chr2_+_172860038 | 0.07 |
ENST00000538974.5
ENST00000540783.5 |
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chrX_+_16650155 | 0.07 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr10_+_116324440 | 0.07 |
ENST00000333254.4
|
CCDC172
|
coiled-coil domain containing 172 |
| chr1_+_167329044 | 0.06 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1 |
| chr1_+_244051275 | 0.06 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr1_-_216423396 | 0.06 |
ENST00000366942.3
ENST00000674083.1 ENST00000307340.8 |
USH2A
|
usherin |
| chr2_+_66435116 | 0.06 |
ENST00000272369.14
ENST00000560281.6 |
MEIS1
|
Meis homeobox 1 |
| chr11_+_121101243 | 0.06 |
ENST00000392793.6
ENST00000642222.1 |
TECTA
|
tectorin alpha |
| chr5_+_51383394 | 0.06 |
ENST00000230658.12
|
ISL1
|
ISL LIM homeobox 1 |
| chr4_+_87799546 | 0.06 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr6_+_110874775 | 0.06 |
ENST00000675380.1
ENST00000368882.8 ENST00000368877.9 ENST00000368885.8 ENST00000672937.2 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr17_-_29005913 | 0.06 |
ENST00000442608.7
ENST00000317338.17 ENST00000335960.10 |
SEZ6
|
seizure related 6 homolog |
| chr14_-_34875348 | 0.06 |
ENST00000360310.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.3 | 0.8 | GO:0072192 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 | 0.5 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.9 | GO:1901963 | regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.1 | 1.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.3 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.3 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.1 | 0.6 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 1.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.1 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0071657 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.5 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 2.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 2.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.5 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.1 | 1.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 2.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0097108 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |