Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HOXA9
Z-value: 0.58
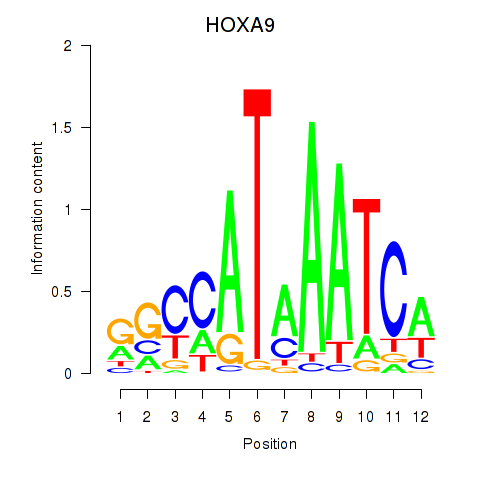
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.19 | HOXA9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg38_v1_chr7_-_27165517_27165545 | 0.32 | 8.3e-02 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_55833186 | 1.78 |
ENST00000361503.8
ENST00000422046.6 |
CES1
|
carboxylesterase 1 |
| chr1_-_152159227 | 1.59 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr12_-_91111460 | 1.54 |
ENST00000266718.5
|
LUM
|
lumican |
| chr2_-_31414694 | 1.41 |
ENST00000379416.4
|
XDH
|
xanthine dehydrogenase |
| chr1_+_24319342 | 1.03 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr4_-_155866277 | 1.01 |
ENST00000537611.3
|
ASIC5
|
acid sensing ion channel subunit family member 5 |
| chr20_+_59628609 | 0.98 |
ENST00000541461.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr6_+_47698538 | 0.91 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr6_+_47698574 | 0.88 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr2_+_142877653 | 0.86 |
ENST00000375773.6
ENST00000409512.5 ENST00000264170.9 ENST00000410015.6 |
KYNU
|
kynureninase |
| chr4_-_48016631 | 0.74 |
ENST00000513178.2
ENST00000514170.7 |
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr12_+_15322257 | 0.73 |
ENST00000674316.1
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr12_+_15322529 | 0.67 |
ENST00000348962.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr21_-_26843063 | 0.66 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr2_+_233195433 | 0.65 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr12_-_52385649 | 0.57 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr11_+_6845683 | 0.55 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr1_+_24319511 | 0.55 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr19_-_48513161 | 0.54 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr6_+_35342535 | 0.51 |
ENST00000360694.8
ENST00000418635.6 ENST00000448077.6 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr2_+_11724333 | 0.50 |
ENST00000425416.6
ENST00000396097.5 |
LPIN1
|
lipin 1 |
| chr3_-_151329539 | 0.48 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr6_-_25832026 | 0.48 |
ENST00000476801.5
ENST00000244527.10 |
SLC17A1
|
solute carrier family 17 member 1 |
| chr6_+_25726767 | 0.47 |
ENST00000274764.5
|
H2BC1
|
H2B clustered histone 1 |
| chr13_-_30306997 | 0.45 |
ENST00000380617.7
ENST00000441394.1 |
KATNAL1
|
katanin catalytic subunit A1 like 1 |
| chr3_-_48089203 | 0.45 |
ENST00000468075.2
ENST00000360240.10 |
MAP4
|
microtubule associated protein 4 |
| chr10_+_100462969 | 0.45 |
ENST00000343737.6
|
WNT8B
|
Wnt family member 8B |
| chr1_+_40988513 | 0.44 |
ENST00000649215.1
|
CTPS1
|
CTP synthase 1 |
| chr4_+_48016764 | 0.44 |
ENST00000295461.10
|
NIPAL1
|
NIPA like domain containing 1 |
| chr2_-_88128049 | 0.44 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chr12_-_10826358 | 0.43 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr3_+_57890011 | 0.42 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr1_+_101237009 | 0.42 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chrX_-_108439472 | 0.42 |
ENST00000372216.8
|
COL4A6
|
collagen type IV alpha 6 chain |
| chr4_-_149815826 | 0.41 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr1_-_204166334 | 0.40 |
ENST00000272190.9
|
REN
|
renin |
| chr7_+_134745460 | 0.40 |
ENST00000436461.6
|
CALD1
|
caldesmon 1 |
| chr18_-_12656716 | 0.39 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr18_+_36544544 | 0.39 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr2_-_189179754 | 0.37 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chrX_-_153321759 | 0.37 |
ENST00000436629.3
|
PNMA6F
|
PNMA family member 6F |
| chr12_+_15322480 | 0.36 |
ENST00000674188.1
ENST00000281171.9 ENST00000543886.6 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr12_+_57460127 | 0.36 |
ENST00000532291.5
ENST00000543426.5 ENST00000546141.5 |
GLI1
|
GLI family zinc finger 1 |
| chr2_+_20447065 | 0.36 |
ENST00000272233.6
|
RHOB
|
ras homolog family member B |
| chr22_+_50170720 | 0.36 |
ENST00000159647.9
ENST00000395842.3 |
PANX2
|
pannexin 2 |
| chr1_-_79006680 | 0.36 |
ENST00000370742.4
ENST00000656841.1 |
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr15_+_67125707 | 0.35 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr5_-_102498913 | 0.35 |
ENST00000513675.1
ENST00000379807.7 ENST00000506729.6 |
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr1_-_152580511 | 0.34 |
ENST00000368787.4
|
LCE3D
|
late cornified envelope 3D |
| chr3_+_172754457 | 0.34 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr11_-_18236795 | 0.34 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr17_-_59151794 | 0.34 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_+_26622800 | 0.34 |
ENST00000396641.6
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr11_-_64778747 | 0.33 |
ENST00000227503.13
ENST00000377394.7 ENST00000334944.9 |
SF1
|
splicing factor 1 |
| chr16_-_88663065 | 0.33 |
ENST00000301012.8
ENST00000569177.5 |
MVD
|
mevalonate diphosphate decarboxylase |
| chr17_-_8210203 | 0.32 |
ENST00000578549.5
ENST00000582368.5 |
AURKB
|
aurora kinase B |
| chr5_+_177304571 | 0.32 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr12_-_13095664 | 0.32 |
ENST00000337630.10
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr12_+_57459782 | 0.32 |
ENST00000228682.7
|
GLI1
|
GLI family zinc finger 1 |
| chr5_+_69189536 | 0.32 |
ENST00000515001.5
ENST00000283006.7 ENST00000502689.1 |
CENPH
|
centromere protein H |
| chr11_-_60184633 | 0.32 |
ENST00000529054.5
ENST00000530839.5 ENST00000426738.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr19_-_11155798 | 0.31 |
ENST00000592540.5
|
SPC24
|
SPC24 component of NDC80 kinetochore complex |
| chr21_+_36135071 | 0.31 |
ENST00000290354.6
|
CBR3
|
carbonyl reductase 3 |
| chr5_-_95081482 | 0.31 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr14_+_67720842 | 0.30 |
ENST00000267502.3
|
RDH12
|
retinol dehydrogenase 12 |
| chr2_+_108378176 | 0.30 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr11_+_57542641 | 0.30 |
ENST00000527972.5
ENST00000399154.3 |
SMTNL1
|
smoothelin like 1 |
| chr19_+_926001 | 0.30 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr9_-_21305313 | 0.29 |
ENST00000610521.2
|
IFNA5
|
interferon alpha 5 |
| chr2_-_70553440 | 0.29 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr5_+_150497772 | 0.29 |
ENST00000523767.5
|
NDST1
|
N-deacetylase and N-sulfotransferase 1 |
| chr6_-_132714045 | 0.29 |
ENST00000367928.5
|
VNN1
|
vanin 1 |
| chr2_-_224947030 | 0.28 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr12_+_80099535 | 0.28 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr15_-_30991415 | 0.28 |
ENST00000563714.5
|
MTMR10
|
myotubularin related protein 10 |
| chr5_-_112419251 | 0.28 |
ENST00000261486.6
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_+_108377947 | 0.28 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr2_-_89143133 | 0.27 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr19_-_41688167 | 0.27 |
ENST00000602225.1
|
CEACAM7
|
CEA cell adhesion molecule 7 |
| chr1_-_198540674 | 0.27 |
ENST00000489986.1
ENST00000367382.6 |
ATP6V1G3
|
ATPase H+ transporting V1 subunit G3 |
| chr6_+_32154131 | 0.27 |
ENST00000375143.6
ENST00000324816.11 ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr12_+_12891554 | 0.27 |
ENST00000014914.6
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A |
| chr5_-_102499008 | 0.27 |
ENST00000389019.7
|
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr4_-_68670648 | 0.27 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr6_+_32154010 | 0.27 |
ENST00000375137.6
|
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr4_-_76023489 | 0.27 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr13_-_30307539 | 0.27 |
ENST00000380615.8
|
KATNAL1
|
katanin catalytic subunit A1 like 1 |
| chr12_-_48004496 | 0.26 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain |
| chr11_+_75768718 | 0.26 |
ENST00000376262.7
|
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr3_-_190449782 | 0.26 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr3_-_197184131 | 0.26 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_112482133 | 0.26 |
ENST00000233336.7
|
TTL
|
tubulin tyrosine ligase |
| chr7_+_54542362 | 0.26 |
ENST00000402613.4
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr14_+_104689588 | 0.25 |
ENST00000330634.11
ENST00000392634.9 ENST00000675482.1 ENST00000398337.8 |
INF2
|
inverted formin 2 |
| chr7_+_142111739 | 0.25 |
ENST00000550469.6
ENST00000477922.3 |
MGAM2
|
maltase-glucoamylase 2 (putative) |
| chr16_+_29679132 | 0.25 |
ENST00000395384.9
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr2_+_89947508 | 0.25 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr1_+_117420597 | 0.24 |
ENST00000449370.6
|
MAN1A2
|
mannosidase alpha class 1A member 2 |
| chr7_+_69599588 | 0.24 |
ENST00000403018.3
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr9_+_74497308 | 0.24 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr6_+_42050876 | 0.24 |
ENST00000465926.5
ENST00000482432.1 |
TAF8
|
TATA-box binding protein associated factor 8 |
| chr1_+_50108856 | 0.23 |
ENST00000650764.1
ENST00000494555.2 ENST00000371824.7 ENST00000371823.8 ENST00000652693.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr6_+_35342614 | 0.23 |
ENST00000337400.6
ENST00000311565.4 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr9_-_21207143 | 0.23 |
ENST00000357374.2
|
IFNA10
|
interferon alpha 10 |
| chr6_+_63571702 | 0.23 |
ENST00000672924.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr9_-_124507382 | 0.23 |
ENST00000373588.9
ENST00000620110.4 |
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr6_-_32407123 | 0.23 |
ENST00000374993.4
ENST00000544175.2 ENST00000454136.7 ENST00000446536.2 |
BTNL2
|
butyrophilin like 2 |
| chr1_+_100719734 | 0.23 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr7_-_80512041 | 0.22 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3 |
| chr4_+_69931066 | 0.22 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr10_+_84173793 | 0.22 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr5_+_127649018 | 0.22 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr2_-_99254281 | 0.22 |
ENST00000409238.5
ENST00000423800.5 |
LYG2
|
lysozyme g2 |
| chr17_-_8118489 | 0.22 |
ENST00000380149.6
ENST00000448843.7 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr6_+_130018565 | 0.21 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr12_-_121039236 | 0.21 |
ENST00000257570.9
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr4_-_163332589 | 0.21 |
ENST00000296533.3
ENST00000509586.5 ENST00000504391.5 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr14_+_94026314 | 0.21 |
ENST00000203664.10
ENST00000553723.1 |
OTUB2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chrX_-_155264471 | 0.21 |
ENST00000369454.4
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr1_+_192158448 | 0.21 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr14_+_22508602 | 0.21 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr11_+_63506046 | 0.21 |
ENST00000674247.1
|
LGALS12
|
galectin 12 |
| chr12_+_106774630 | 0.21 |
ENST00000392839.6
ENST00000548914.5 ENST00000355478.6 ENST00000552619.1 ENST00000549643.5 ENST00000392837.9 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr12_-_91179472 | 0.21 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr8_+_7836433 | 0.21 |
ENST00000314265.3
|
DEFB104A
|
defensin beta 104A |
| chrX_-_132219473 | 0.20 |
ENST00000620646.4
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr9_-_21166660 | 0.20 |
ENST00000380225.1
|
IFNA21
|
interferon alpha 21 |
| chr7_+_120988683 | 0.20 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chrX_-_132219439 | 0.20 |
ENST00000370874.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr12_+_18262730 | 0.20 |
ENST00000675017.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr19_-_10309783 | 0.20 |
ENST00000403352.1
ENST00000403903.7 |
ZGLP1
|
zinc finger GATA like protein 1 |
| chr7_-_36724380 | 0.20 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr19_-_6375849 | 0.19 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr16_+_70579867 | 0.19 |
ENST00000429149.6
|
IL34
|
interleukin 34 |
| chr3_+_63443076 | 0.19 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr6_-_116545658 | 0.19 |
ENST00000368602.4
|
TRAPPC3L
|
trafficking protein particle complex 3 like |
| chr9_-_21187671 | 0.19 |
ENST00000421715.2
|
IFNA4
|
interferon alpha 4 |
| chr8_+_12108172 | 0.19 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D |
| chr2_+_190137760 | 0.19 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr11_+_59755427 | 0.19 |
ENST00000529177.5
|
STX3
|
syntaxin 3 |
| chr12_-_121039204 | 0.19 |
ENST00000620239.5
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr17_-_31297231 | 0.19 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr12_-_11134644 | 0.19 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chrX_-_111270474 | 0.19 |
ENST00000324068.2
|
CAPN6
|
calpain 6 |
| chr8_+_38404363 | 0.19 |
ENST00000527175.1
|
LETM2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr5_-_83720813 | 0.19 |
ENST00000515590.1
ENST00000274341.9 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr12_-_121039156 | 0.19 |
ENST00000339275.10
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr9_-_92482499 | 0.19 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr17_-_39197652 | 0.18 |
ENST00000394303.8
|
CACNB1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr21_-_18403754 | 0.18 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15 |
| chr15_+_51829644 | 0.18 |
ENST00000308580.12
|
TMOD3
|
tropomodulin 3 |
| chr11_-_71821548 | 0.18 |
ENST00000525199.1
|
ZNF705E
|
zinc finger protein 705E |
| chr10_+_122560679 | 0.18 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr17_+_75667390 | 0.18 |
ENST00000583536.5
|
SAP30BP
|
SAP30 binding protein |
| chr12_+_31073849 | 0.18 |
ENST00000228264.10
ENST00000542838.6 ENST00000438391.6 ENST00000415475.6 ENST00000545668.5 ENST00000350437.8 |
DDX11
|
DEAD/H-box helicase 11 |
| chr3_-_195583931 | 0.17 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr9_-_92482350 | 0.17 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr6_-_29431967 | 0.17 |
ENST00000377154.1
ENST00000641152.2 |
OR5V1
OR11A1
|
olfactory receptor family 5 subfamily V member 1 olfactory receptor family 11 subfamily A member 1 |
| chr7_-_106285898 | 0.17 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr5_-_175961324 | 0.17 |
ENST00000432305.6
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr19_-_41688258 | 0.17 |
ENST00000401731.6
ENST00000006724.7 |
CEACAM7
|
CEA cell adhesion molecule 7 |
| chr15_+_21579912 | 0.17 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr10_+_122560639 | 0.17 |
ENST00000344338.7
ENST00000330163.8 ENST00000652446.2 ENST00000666315.1 ENST00000368955.7 ENST00000368909.7 ENST00000368956.6 ENST00000619379.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr22_-_33058368 | 0.17 |
ENST00000358763.7
|
SYN3
|
synapsin III |
| chr10_+_122560751 | 0.17 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr4_+_113049479 | 0.17 |
ENST00000671727.1
ENST00000671762.1 ENST00000672366.1 ENST00000672502.1 ENST00000672045.1 ENST00000672251.1 ENST00000672854.1 |
ANK2
|
ankyrin 2 |
| chr6_+_63563448 | 0.16 |
ENST00000673199.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr11_-_55936400 | 0.16 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr12_-_54384687 | 0.16 |
ENST00000550120.1
ENST00000547210.5 ENST00000394313.7 |
ZNF385A
|
zinc finger protein 385A |
| chr6_-_27807916 | 0.16 |
ENST00000377401.3
|
H2BC13
|
H2B clustered histone 13 |
| chrX_-_120878924 | 0.16 |
ENST00000613352.1
|
ENSG00000278646.1
|
novel protein similar to cancer/testis antigen family 47, member A12 (CT47A12) |
| chr11_+_60842095 | 0.16 |
ENST00000227520.10
|
CCDC86
|
coiled-coil domain containing 86 |
| chr10_+_89332484 | 0.16 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr5_-_149379286 | 0.16 |
ENST00000261796.4
|
IL17B
|
interleukin 17B |
| chr11_-_105023136 | 0.16 |
ENST00000526056.5
ENST00000531367.5 ENST00000456094.1 ENST00000444749.6 ENST00000393141.6 ENST00000418434.5 ENST00000260315.8 |
CASP5
|
caspase 5 |
| chr7_-_16465728 | 0.16 |
ENST00000307068.5
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr7_-_14841267 | 0.16 |
ENST00000406247.7
ENST00000399322.7 |
DGKB
|
diacylglycerol kinase beta |
| chr22_+_22906342 | 0.16 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr2_-_2324642 | 0.16 |
ENST00000650485.1
ENST00000649207.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr6_-_52087569 | 0.16 |
ENST00000340994.4
ENST00000371117.8 |
PKHD1
|
PKHD1 ciliary IPT domain containing fibrocystin/polyductin |
| chr17_+_62370218 | 0.16 |
ENST00000450662.7
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr17_-_41572052 | 0.16 |
ENST00000588431.1
ENST00000246662.9 |
KRT9
|
keratin 9 |
| chr10_-_88952763 | 0.16 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle |
| chr1_-_119811458 | 0.16 |
ENST00000256585.10
ENST00000354219.5 ENST00000369401.4 |
REG4
|
regenerating family member 4 |
| chr14_+_22040576 | 0.15 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr6_-_32154326 | 0.15 |
ENST00000475826.1
ENST00000485392.5 ENST00000494332.5 ENST00000498575.1 ENST00000428778.5 |
ENSG00000284954.1
ENSG00000285085.1
|
novel transcript novel protein |
| chr10_-_44978789 | 0.15 |
ENST00000448778.1
ENST00000298295.4 |
DEPP1
|
DEPP1 autophagy regulator |
| chr2_+_203867764 | 0.15 |
ENST00000648405.2
|
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr15_-_22160868 | 0.15 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr11_-_104968533 | 0.15 |
ENST00000444739.7
|
CASP4
|
caspase 4 |
| chr20_-_50115935 | 0.15 |
ENST00000340309.7
ENST00000415862.6 ENST00000371677.7 |
UBE2V1
|
ubiquitin conjugating enzyme E2 V1 |
| chr1_+_218345326 | 0.15 |
ENST00000366930.9
|
TGFB2
|
transforming growth factor beta 2 |
| chr3_-_173141227 | 0.15 |
ENST00000351008.4
|
SPATA16
|
spermatogenesis associated 16 |
| chr5_-_132011580 | 0.15 |
ENST00000651250.1
ENST00000434099.6 ENST00000296869.9 ENST00000651356.1 ENST00000651883.2 |
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
| chr19_-_47471886 | 0.15 |
ENST00000236877.11
ENST00000597014.1 |
SLC8A2
|
solute carrier family 8 member A2 |
| chr9_-_21228222 | 0.15 |
ENST00000413767.2
|
IFNA17
|
interferon alpha 17 |
| chr6_+_25279359 | 0.15 |
ENST00000329474.7
|
CARMIL1
|
capping protein regulator and myosin 1 linker 1 |
| chr14_+_22112280 | 0.15 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr21_-_30480364 | 0.15 |
ENST00000390689.3
|
KRTAP19-1
|
keratin associated protein 19-1 |
| chr8_-_13514744 | 0.15 |
ENST00000316609.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.4 | 1.8 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.4 | 1.8 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 0.7 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 0.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.7 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 1.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.1 | 1.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.3 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.7 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 1.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.3 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.4 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.1 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.4 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:0090131 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.0 | 0.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 2.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0009631 | cold acclimation(GO:0009631) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.3 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.3 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 2.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0097025 | myelin sheath abaxonal region(GO:0035748) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 1.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 1.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.9 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.5 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 0.5 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.3 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 1.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.7 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.9 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |