Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HOXB3
Z-value: 0.41
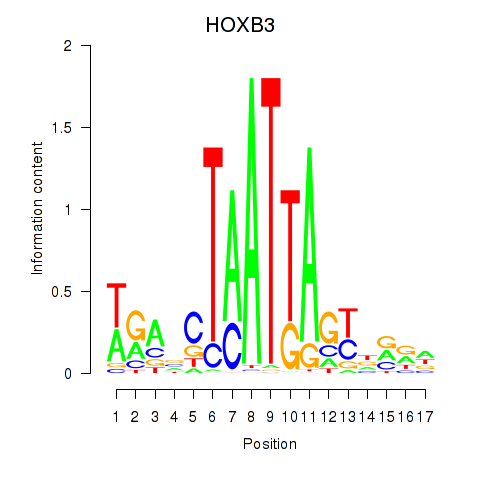
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.12 | HOXB3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB3 | hg38_v1_chr17_-_48590231_48590258 | 0.05 | 7.9e-01 | Click! |
Activity profile of HOXB3 motif
Sorted Z-values of HOXB3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_152514474 | 1.37 |
ENST00000368790.4
|
CRCT1
|
cysteine rich C-terminal 1 |
| chr22_+_30396991 | 0.78 |
ENST00000617837.4
ENST00000615189.5 ENST00000405717.7 ENST00000402592.7 |
SEC14L2
|
SEC14 like lipid binding 2 |
| chr4_+_85604146 | 0.74 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_-_22853320 | 0.66 |
ENST00000504376.6
ENST00000382254.6 |
CDH12
|
cadherin 12 |
| chr3_+_122055355 | 0.61 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr2_+_113058637 | 0.57 |
ENST00000346807.7
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr22_+_44026283 | 0.53 |
ENST00000619710.4
|
PARVB
|
parvin beta |
| chr2_+_113059194 | 0.51 |
ENST00000393200.7
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr4_-_149815826 | 0.51 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr11_-_107858777 | 0.49 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chrX_+_139530730 | 0.49 |
ENST00000218099.7
|
F9
|
coagulation factor IX |
| chr12_+_15546344 | 0.48 |
ENST00000674388.1
ENST00000542557.5 ENST00000445537.6 ENST00000544244.5 ENST00000442921.7 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr3_-_52535006 | 0.47 |
ENST00000307076.8
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr17_+_42458844 | 0.47 |
ENST00000393829.6
ENST00000537728.5 ENST00000343619.9 ENST00000264649.10 ENST00000585525.5 ENST00000544137.5 ENST00000589727.5 ENST00000587824.5 |
ATP6V0A1
|
ATPase H+ transporting V0 subunit a1 |
| chr11_+_33039996 | 0.46 |
ENST00000432887.5
ENST00000528898.1 ENST00000531632.6 |
TCP11L1
|
t-complex 11 like 1 |
| chr8_+_27311471 | 0.44 |
ENST00000397501.5
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr9_+_72577369 | 0.43 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr12_-_10826358 | 0.42 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr4_+_143433491 | 0.41 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr11_-_124320197 | 0.41 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr3_+_136819069 | 0.39 |
ENST00000393079.3
ENST00000446465.3 |
SLC35G2
|
solute carrier family 35 member G2 |
| chr5_+_151259793 | 0.39 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr8_-_30812867 | 0.38 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr2_+_44275457 | 0.38 |
ENST00000611973.4
ENST00000409387.5 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr12_-_122703346 | 0.37 |
ENST00000328880.6
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr22_-_29838227 | 0.35 |
ENST00000307790.8
ENST00000397771.6 ENST00000542393.5 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr3_-_185821092 | 0.34 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr12_+_20695553 | 0.33 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr21_+_42403874 | 0.32 |
ENST00000319294.11
ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr9_+_133061981 | 0.32 |
ENST00000372080.8
|
CEL
|
carboxyl ester lipase |
| chrX_+_111876177 | 0.32 |
ENST00000635763.2
|
TRPC5OS
|
TRPC5 opposite strand |
| chr8_-_48921419 | 0.32 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr12_-_122716790 | 0.32 |
ENST00000528880.3
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr9_+_122371036 | 0.31 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr8_-_13276491 | 0.31 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr7_-_101217569 | 0.31 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr9_+_72577788 | 0.31 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr4_+_122378966 | 0.30 |
ENST00000446706.5
ENST00000296513.7 |
ADAD1
|
adenosine deaminase domain containing 1 |
| chr11_-_118264445 | 0.30 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero like 2 |
| chr6_-_110179995 | 0.30 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr12_-_52652207 | 0.29 |
ENST00000309680.4
|
KRT2
|
keratin 2 |
| chr7_-_100167284 | 0.28 |
ENST00000413800.5
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4 |
| chr8_+_7926337 | 0.27 |
ENST00000400120.3
|
ZNF705B
|
zinc finger protein 705B |
| chr14_-_106038355 | 0.27 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr15_+_71810539 | 0.27 |
ENST00000617575.5
ENST00000621098.1 |
NR2E3
|
nuclear receptor subfamily 2 group E member 3 |
| chr2_+_90038848 | 0.27 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr9_+_131289685 | 0.27 |
ENST00000372264.4
|
PLPP7
|
phospholipid phosphatase 7 (inactive) |
| chr6_-_116545658 | 0.27 |
ENST00000368602.4
|
TRAPPC3L
|
trafficking protein particle complex 3 like |
| chr2_+_27275429 | 0.26 |
ENST00000420191.5
ENST00000296097.8 |
DNAJC5G
|
DnaJ heat shock protein family (Hsp40) member C5 gamma |
| chr2_+_48314637 | 0.26 |
ENST00000413569.5
ENST00000340553.8 |
FOXN2
|
forkhead box N2 |
| chr6_+_160121859 | 0.26 |
ENST00000324965.8
ENST00000457470.6 |
SLC22A1
|
solute carrier family 22 member 1 |
| chr12_+_22625182 | 0.26 |
ENST00000538218.2
|
ETNK1
|
ethanolamine kinase 1 |
| chr11_+_65787056 | 0.25 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr6_+_54018910 | 0.25 |
ENST00000514921.5
ENST00000274897.9 ENST00000370877.6 |
MLIP
|
muscular LMNA interacting protein |
| chr6_+_160121809 | 0.25 |
ENST00000366963.9
|
SLC22A1
|
solute carrier family 22 member 1 |
| chr8_+_22275309 | 0.25 |
ENST00000356766.11
ENST00000521356.5 |
PIWIL2
|
piwi like RNA-mediated gene silencing 2 |
| chr8_+_12104389 | 0.25 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr14_-_24242320 | 0.24 |
ENST00000557921.2
|
TINF2
|
TERF1 interacting nuclear factor 2 |
| chr2_+_29113989 | 0.23 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr1_-_66801276 | 0.23 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr20_-_56525925 | 0.23 |
ENST00000243913.8
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr12_-_7936177 | 0.23 |
ENST00000544291.1
ENST00000075120.12 |
SLC2A3
|
solute carrier family 2 member 3 |
| chr4_+_25160631 | 0.23 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chr1_+_15764419 | 0.22 |
ENST00000441801.6
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr6_+_26204552 | 0.22 |
ENST00000615164.2
|
H4C5
|
H4 clustered histone 5 |
| chr7_-_111392915 | 0.22 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr19_+_7830188 | 0.22 |
ENST00000270530.8
|
EVI5L
|
ecotropic viral integration site 5 like |
| chr19_+_49766962 | 0.22 |
ENST00000354293.10
ENST00000359032.9 |
AP2A1
|
adaptor related protein complex 2 subunit alpha 1 |
| chr6_-_127459364 | 0.21 |
ENST00000487331.2
ENST00000483725.8 |
KIAA0408
|
KIAA0408 |
| chr14_+_56117702 | 0.21 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr21_-_26051023 | 0.21 |
ENST00000415997.1
|
APP
|
amyloid beta precursor protein |
| chr14_+_32329341 | 0.21 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr7_+_120988683 | 0.21 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chrX_+_43656289 | 0.21 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr19_-_5286163 | 0.21 |
ENST00000592099.5
ENST00000588012.5 ENST00000262963.10 ENST00000587303.5 ENST00000590509.5 |
PTPRS
|
protein tyrosine phosphatase receptor type S |
| chr4_-_142846275 | 0.20 |
ENST00000513000.5
ENST00000509777.5 ENST00000503927.5 |
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr8_+_144148027 | 0.20 |
ENST00000423230.6
|
MROH1
|
maestro heat like repeat family member 1 |
| chr10_-_133565542 | 0.20 |
ENST00000303903.10
ENST00000343131.7 |
SYCE1
|
synaptonemal complex central element protein 1 |
| chr6_+_130018565 | 0.20 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr9_+_122371014 | 0.20 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr19_-_3557563 | 0.19 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr1_+_207053229 | 0.19 |
ENST00000367080.8
ENST00000367079.3 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr19_-_17255448 | 0.19 |
ENST00000594059.1
|
ENSG00000269095.1
|
novel protein |
| chr15_-_55270280 | 0.19 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_21910853 | 0.19 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr18_-_46964408 | 0.18 |
ENST00000676383.1
|
ENSG00000288616.1
|
elongin A3 family member D |
| chr2_+_28395511 | 0.18 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr18_+_58221535 | 0.18 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chrX_-_108736556 | 0.18 |
ENST00000372129.4
|
IRS4
|
insulin receptor substrate 4 |
| chr15_+_78438279 | 0.18 |
ENST00000560440.5
|
IREB2
|
iron responsive element binding protein 2 |
| chr2_+_88885397 | 0.18 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr12_+_56128217 | 0.18 |
ENST00000267113.4
ENST00000394048.10 |
ESYT1
|
extended synaptotagmin 1 |
| chr8_-_7385558 | 0.17 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr12_+_8157034 | 0.17 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr5_-_126595237 | 0.17 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr15_-_55270874 | 0.17 |
ENST00000567380.5
ENST00000565972.5 ENST00000569493.5 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr9_+_121567057 | 0.17 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr14_+_94174284 | 0.17 |
ENST00000304338.8
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr19_-_14848922 | 0.16 |
ENST00000641129.1
|
OR7A10
|
olfactory receptor family 7 subfamily A member 10 |
| chr18_-_24311495 | 0.16 |
ENST00000357041.8
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr6_-_110815408 | 0.16 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19 |
| chr12_-_11395556 | 0.16 |
ENST00000565533.1
ENST00000389362.6 ENST00000546254.3 |
PRB2
PRB1
|
proline rich protein BstNI subfamily 2 proline rich protein BstNI subfamily 1 |
| chr16_+_6019016 | 0.16 |
ENST00000550418.6
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr2_-_101308681 | 0.16 |
ENST00000295317.4
|
RNF149
|
ring finger protein 149 |
| chr6_+_47698538 | 0.16 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr12_+_119668109 | 0.16 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr14_+_94174334 | 0.16 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr22_+_22811737 | 0.15 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr17_+_50547089 | 0.15 |
ENST00000619622.4
ENST00000356488.8 ENST00000006658.11 |
SPATA20
|
spermatogenesis associated 20 |
| chr16_+_11965193 | 0.15 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr2_+_233636445 | 0.15 |
ENST00000344644.9
|
UGT1A10
|
UDP glucuronosyltransferase family 1 member A10 |
| chr5_+_149357999 | 0.15 |
ENST00000274569.9
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr1_-_207052980 | 0.15 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr6_+_47698574 | 0.14 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr16_+_11965234 | 0.14 |
ENST00000562385.1
|
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr17_+_48892761 | 0.14 |
ENST00000355938.9
ENST00000393366.7 ENST00000503641.5 ENST00000514808.5 ENST00000506855.1 |
ATP5MC1
|
ATP synthase membrane subunit c locus 1 |
| chr22_-_28711931 | 0.14 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr2_+_64988469 | 0.14 |
ENST00000531327.5
|
SLC1A4
|
solute carrier family 1 member 4 |
| chr2_-_199851114 | 0.14 |
ENST00000420128.5
ENST00000416668.5 ENST00000622774.2 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr17_-_59151794 | 0.14 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr16_+_6019071 | 0.14 |
ENST00000547605.5
ENST00000553186.5 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr3_+_114294020 | 0.14 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr11_+_95789965 | 0.14 |
ENST00000537677.5
|
CEP57
|
centrosomal protein 57 |
| chr12_-_118359639 | 0.13 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr6_-_110815152 | 0.13 |
ENST00000413605.6
|
CDK19
|
cyclin dependent kinase 19 |
| chr19_+_49487510 | 0.13 |
ENST00000679106.1
ENST00000621674.4 ENST00000391857.9 ENST00000678510.1 ENST00000467825.2 |
RPL13A
|
ribosomal protein L13a |
| chr2_+_233729042 | 0.13 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr18_+_63887698 | 0.13 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr13_+_45464901 | 0.13 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr2_-_165203870 | 0.13 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr15_-_101294905 | 0.13 |
ENST00000560496.5
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr11_+_24496988 | 0.13 |
ENST00000336930.11
|
LUZP2
|
leucine zipper protein 2 |
| chrX_-_21658324 | 0.13 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr14_-_94476240 | 0.12 |
ENST00000424550.6
ENST00000337425.10 ENST00000674164.1 ENST00000546329.2 |
SERPINA9
|
serpin family A member 9 |
| chr19_+_48445961 | 0.12 |
ENST00000253237.10
|
GRWD1
|
glutamate rich WD repeat containing 1 |
| chrX_+_86714623 | 0.12 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr12_-_91058016 | 0.12 |
ENST00000266719.4
|
KERA
|
keratocan |
| chr2_+_142877653 | 0.12 |
ENST00000375773.6
ENST00000409512.5 ENST00000264170.9 ENST00000410015.6 |
KYNU
|
kynureninase |
| chr1_-_51331315 | 0.12 |
ENST00000262676.9
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr17_-_46361718 | 0.12 |
ENST00000575960.5
ENST00000575698.5 ENST00000571246.5 ENST00000434041.6 ENST00000656849.1 ENST00000570618.5 ENST00000450673.4 ENST00000622877.4 |
ARL17B
|
ADP ribosylation factor like GTPase 17B |
| chr6_-_169250825 | 0.12 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr4_-_68245683 | 0.12 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B |
| chr8_+_7836433 | 0.11 |
ENST00000314265.3
|
DEFB104A
|
defensin beta 104A |
| chr1_-_197146620 | 0.11 |
ENST00000367409.9
ENST00000680265.1 |
ASPM
|
assembly factor for spindle microtubules |
| chr22_-_32255344 | 0.11 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chr19_+_9185594 | 0.11 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr22_-_28712136 | 0.11 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2 |
| chr21_+_46286623 | 0.11 |
ENST00000397691.1
|
YBEY
|
ybeY metalloendoribonuclease |
| chr1_-_89175997 | 0.11 |
ENST00000294671.3
ENST00000650452.1 |
GBP7
|
guanylate binding protein 7 |
| chr4_-_110641920 | 0.11 |
ENST00000354925.6
ENST00000511990.1 ENST00000613094.4 ENST00000614423.4 ENST00000616641.4 ENST00000511837.5 |
PITX2
|
paired like homeodomain 2 |
| chr2_-_88861258 | 0.11 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr3_-_165078480 | 0.11 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr2_+_86942118 | 0.10 |
ENST00000641458.2
|
RGPD1
|
RANBP2 like and GRIP domain containing 1 |
| chr11_+_95790459 | 0.10 |
ENST00000325486.9
ENST00000325542.10 ENST00000544522.5 ENST00000541365.5 |
CEP57
|
centrosomal protein 57 |
| chr1_-_153150884 | 0.10 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr12_-_10453330 | 0.10 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr6_+_127577168 | 0.10 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr1_+_186296267 | 0.10 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr7_-_33062750 | 0.10 |
ENST00000610140.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr3_+_43690880 | 0.10 |
ENST00000458276.7
|
ABHD5
|
abhydrolase domain containing 5, lysophosphatidic acid acyltransferase |
| chr14_-_24242600 | 0.10 |
ENST00000646753.1
ENST00000558566.1 ENST00000267415.12 ENST00000559019.1 ENST00000399423.8 ENST00000626689.2 |
TINF2
|
TERF1 interacting nuclear factor 2 |
| chrX_+_13689116 | 0.10 |
ENST00000464506.2
ENST00000243325.6 |
RAB9A
|
RAB9A, member RAS oncogene family |
| chr1_+_34792990 | 0.10 |
ENST00000450137.1
ENST00000342280.5 |
GJA4
|
gap junction protein alpha 4 |
| chr7_+_101218146 | 0.10 |
ENST00000305105.3
|
ZNHIT1
|
zinc finger HIT-type containing 1 |
| chrX_+_91434829 | 0.10 |
ENST00000312600.4
|
PABPC5
|
poly(A) binding protein cytoplasmic 5 |
| chr8_-_140800535 | 0.10 |
ENST00000521986.5
ENST00000523539.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr6_+_63571702 | 0.09 |
ENST00000672924.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr17_-_46579651 | 0.09 |
ENST00000573185.5
ENST00000570550.5 ENST00000445552.6 ENST00000329240.10 ENST00000622488.6 ENST00000336125.6 |
ARL17A
|
ADP ribosylation factor like GTPase 17A |
| chr12_+_107685759 | 0.09 |
ENST00000412830.8
ENST00000547995.5 |
PWP1
|
PWP1 homolog, endonuclein |
| chrX_+_37349287 | 0.09 |
ENST00000466533.5
ENST00000542554.5 ENST00000543642.5 ENST00000484460.5 ENST00000378628.9 ENST00000449135.6 ENST00000463135.1 ENST00000465127.1 |
PRRG1
ENSG00000250349.3
|
proline rich and Gla domain 1 novel proline rich Gla (G-carboxyglutamic acid) 1 (PRRG1) and tetraspanin 7 (TSPAN7) protein |
| chr9_+_12775012 | 0.09 |
ENST00000319264.4
|
LURAP1L
|
leucine rich adaptor protein 1 like |
| chr1_+_197268222 | 0.09 |
ENST00000367400.8
ENST00000638467.1 ENST00000367399.6 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr17_+_50548041 | 0.09 |
ENST00000634597.1
|
SPATA20
|
spermatogenesis associated 20 |
| chr11_-_13496018 | 0.09 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr1_-_201171545 | 0.09 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr2_+_11542662 | 0.09 |
ENST00000389825.7
ENST00000381483.6 |
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr11_+_24497155 | 0.09 |
ENST00000529015.5
ENST00000533227.5 |
LUZP2
|
leucine zipper protein 2 |
| chr5_+_132873660 | 0.09 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr2_+_233671879 | 0.09 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr1_+_197268204 | 0.09 |
ENST00000535699.5
ENST00000538660.5 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr1_+_161721563 | 0.09 |
ENST00000367948.6
|
FCRLB
|
Fc receptor like B |
| chr19_+_54137740 | 0.09 |
ENST00000644245.1
ENST00000646002.1 ENST00000221232.11 ENST00000440571.6 ENST00000617930.2 |
CNOT3
|
CCR4-NOT transcription complex subunit 3 |
| chr3_+_122183664 | 0.09 |
ENST00000639785.2
|
CASR
|
calcium sensing receptor |
| chr19_+_49527988 | 0.09 |
ENST00000270645.8
|
RCN3
|
reticulocalbin 3 |
| chr4_+_150078426 | 0.09 |
ENST00000296550.12
|
DCLK2
|
doublecortin like kinase 2 |
| chrX_-_103502853 | 0.08 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr5_-_34043205 | 0.08 |
ENST00000382065.8
ENST00000231338.7 |
C1QTNF3
|
C1q and TNF related 3 |
| chr8_-_42768602 | 0.08 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr12_-_50249883 | 0.08 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr3_-_151329539 | 0.08 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr14_+_21852457 | 0.08 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr14_-_54441325 | 0.08 |
ENST00000556113.1
ENST00000553660.5 ENST00000216416.9 ENST00000395573.8 ENST00000557690.5 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr2_+_186694007 | 0.08 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr10_-_114144599 | 0.08 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr2_+_137964279 | 0.08 |
ENST00000329366.8
|
HNMT
|
histamine N-methyltransferase |
| chr19_-_3985451 | 0.08 |
ENST00000309311.7
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr2_+_165239388 | 0.08 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr6_+_24667026 | 0.08 |
ENST00000537591.5
ENST00000230048.5 |
ACOT13
|
acyl-CoA thioesterase 13 |
| chr3_+_98168700 | 0.08 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr6_-_118935114 | 0.08 |
ENST00000316068.7
|
MCM9
|
minichromosome maintenance 9 homologous recombination repair factor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043016 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 0.4 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 1.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.1 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.3 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.3 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0051563 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0032499 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) cellular response to muramyl dipeptide(GO:0071225) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.4 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.4 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.1 | 0.5 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0050211 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |