Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HOXB4_LHX9
Z-value: 0.84
Transcription factors associated with HOXB4_LHX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
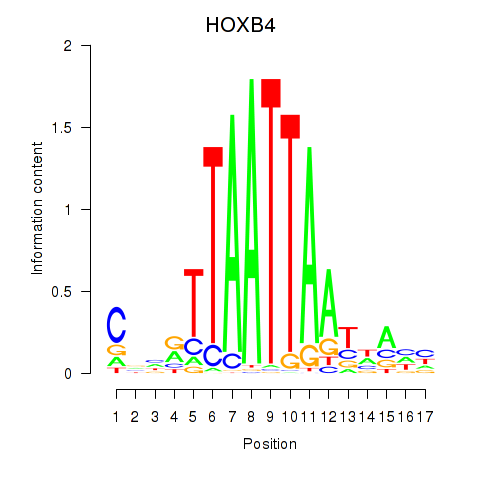
HOXB4
|
ENSG00000182742.6 | HOXB4 |
|
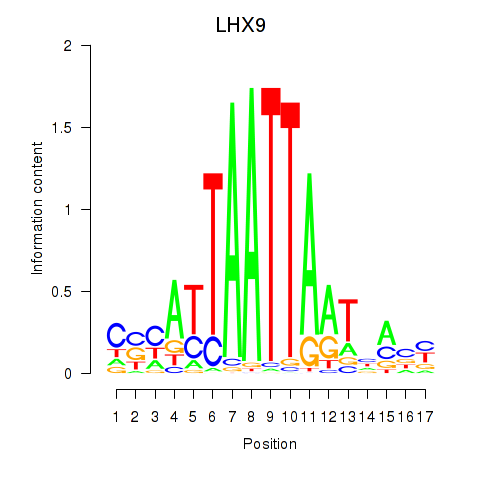
LHX9
|
ENSG00000143355.16 | LHX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX9 | hg38_v1_chr1_+_197912462_197912505 | 0.49 | 6.6e-03 | Click! |
| HOXB4 | hg38_v1_chr17_-_48578341_48578356 | 0.39 | 3.2e-02 | Click! |
Activity profile of HOXB4_LHX9 motif
Sorted Z-values of HOXB4_LHX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB4_LHX9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_72577939 | 4.48 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr10_-_104085847 | 3.97 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr2_+_102337148 | 3.59 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr11_-_107858777 | 3.29 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr12_-_10826358 | 3.05 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr2_-_160200289 | 2.97 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6 |
| chr9_+_122371036 | 2.72 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_72577369 | 2.68 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr2_-_160200251 | 2.62 |
ENST00000428609.6
ENST00000409967.6 ENST00000283249.7 |
ITGB6
|
integrin subunit beta 6 |
| chr17_+_59155726 | 2.58 |
ENST00000578777.5
ENST00000577457.1 ENST00000582995.5 ENST00000262293.9 ENST00000614081.1 |
PRR11
|
proline rich 11 |
| chr2_-_160200310 | 2.37 |
ENST00000620391.4
|
ITGB6
|
integrin subunit beta 6 |
| chr8_-_124565699 | 2.29 |
ENST00000519168.5
|
MTSS1
|
MTSS I-BAR domain containing 1 |
| chr6_+_130018565 | 2.29 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr11_+_33039996 | 2.25 |
ENST00000432887.5
ENST00000528898.1 ENST00000531632.6 |
TCP11L1
|
t-complex 11 like 1 |
| chrX_+_43656289 | 2.23 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr4_-_39032343 | 2.17 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr2_+_90038848 | 2.05 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr7_+_134843884 | 2.05 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr18_+_31447732 | 1.91 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr12_+_119668109 | 1.76 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr9_+_122371014 | 1.74 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr2_+_90172802 | 1.70 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr5_-_140346596 | 1.69 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr11_-_122116215 | 1.64 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr14_-_67412112 | 1.64 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr5_-_126595237 | 1.58 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr12_-_9869345 | 1.52 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr21_-_26843012 | 1.47 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr4_-_159035226 | 1.45 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr2_+_101839815 | 1.38 |
ENST00000421882.5
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr9_-_74952904 | 1.37 |
ENST00000376854.6
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr9_+_72577788 | 1.37 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr21_-_26843063 | 1.37 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr4_+_85604146 | 1.32 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr15_+_21579912 | 1.30 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr14_+_56117702 | 1.28 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr12_+_26195647 | 1.27 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr12_-_21910853 | 1.22 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr5_+_90474879 | 1.21 |
ENST00000504930.5
ENST00000514483.5 |
POLR3G
|
RNA polymerase III subunit G |
| chr15_+_22015233 | 1.21 |
ENST00000639059.1
ENST00000640156.1 |
ENSG00000285472.1
ENSG00000284500.3
|
novel protein novel transcript |
| chr11_-_117316230 | 1.21 |
ENST00000313005.11
ENST00000528053.5 |
BACE1
|
beta-secretase 1 |
| chr12_+_26195313 | 1.20 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr9_+_122370523 | 1.20 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr18_+_23873000 | 1.20 |
ENST00000269217.11
ENST00000587184.5 |
LAMA3
|
laminin subunit alpha 3 |
| chr5_+_90474848 | 1.19 |
ENST00000651687.1
|
POLR3G
|
RNA polymerase III subunit G |
| chr14_+_103385450 | 1.19 |
ENST00000416682.6
|
MARK3
|
microtubule affinity regulating kinase 3 |
| chr2_+_181986015 | 1.16 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C |
| chr9_+_121567057 | 1.14 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr20_-_57690624 | 1.08 |
ENST00000414037.5
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_-_197146620 | 1.08 |
ENST00000367409.9
ENST00000680265.1 |
ASPM
|
assembly factor for spindle microtubules |
| chr3_-_185821092 | 1.07 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chrX_-_101407893 | 1.05 |
ENST00000676156.1
ENST00000675592.1 ENST00000674634.2 ENST00000649178.1 ENST00000218516.4 |
GLA
|
galactosidase alpha |
| chr4_-_39977836 | 1.03 |
ENST00000303538.13
ENST00000503396.5 |
PDS5A
|
PDS5 cohesin associated factor A |
| chr7_+_107583919 | 1.03 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr6_-_110179995 | 0.99 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr19_+_53962925 | 0.99 |
ENST00000270458.4
|
CACNG8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr11_+_35180279 | 0.97 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr14_-_106038355 | 0.95 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr17_+_35587478 | 0.94 |
ENST00000618940.4
|
AP2B1
|
adaptor related protein complex 2 subunit beta 1 |
| chr6_+_113857333 | 0.92 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr14_+_24171853 | 0.90 |
ENST00000620473.4
ENST00000557806.5 ENST00000611366.5 |
REC8
|
REC8 meiotic recombination protein |
| chr15_+_67125707 | 0.90 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr2_-_89085787 | 0.87 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr7_+_134745460 | 0.86 |
ENST00000436461.6
|
CALD1
|
caldesmon 1 |
| chr2_+_90234809 | 0.86 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr3_-_142029108 | 0.86 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr12_+_26195543 | 0.85 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr6_-_82247697 | 0.85 |
ENST00000306270.12
ENST00000610980.4 |
IBTK
|
inhibitor of Bruton tyrosine kinase |
| chr1_+_152811971 | 0.84 |
ENST00000360090.4
|
LCE1B
|
late cornified envelope 1B |
| chr1_-_13116854 | 0.83 |
ENST00000621994.3
|
HNRNPCL2
|
heterogeneous nuclear ribonucleoprotein C like 2 |
| chr20_-_52105644 | 0.83 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chrX_-_72239022 | 0.81 |
ENST00000373657.2
ENST00000334463.4 |
ERCC6L
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr2_+_181985846 | 0.81 |
ENST00000682840.1
ENST00000409137.7 ENST00000280295.7 |
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C |
| chr4_+_168497066 | 0.79 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_-_103076688 | 0.79 |
ENST00000394785.9
|
SLC9B2
|
solute carrier family 9 member B2 |
| chr1_-_197146688 | 0.78 |
ENST00000294732.11
|
ASPM
|
assembly factor for spindle microtubules |
| chr11_-_69819410 | 0.78 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr11_+_55811367 | 0.76 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr6_+_29111560 | 0.76 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr8_-_30812867 | 0.75 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr4_+_70226116 | 0.74 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr7_-_87713287 | 0.73 |
ENST00000416177.1
ENST00000265724.8 ENST00000543898.5 |
ABCB1
|
ATP binding cassette subfamily B member 1 |
| chr2_+_87338511 | 0.73 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr4_+_168497044 | 0.73 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_25160631 | 0.72 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chr14_+_103385374 | 0.72 |
ENST00000678179.1
ENST00000676938.1 ENST00000678619.1 ENST00000440884.7 ENST00000560417.6 ENST00000679330.1 ENST00000556744.2 ENST00000676897.1 ENST00000677560.1 ENST00000561314.6 ENST00000677829.1 ENST00000677133.1 ENST00000676645.1 ENST00000678175.1 ENST00000429436.7 ENST00000677360.1 ENST00000678237.1 ENST00000677347.1 ENST00000677432.1 |
MARK3
|
microtubule affinity regulating kinase 3 |
| chr4_-_119322128 | 0.72 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chrX_-_18672101 | 0.71 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr6_+_47781982 | 0.71 |
ENST00000489301.6
ENST00000638973.1 ENST00000371211.6 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr2_-_88979016 | 0.71 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr5_-_16916400 | 0.71 |
ENST00000513882.5
|
MYO10
|
myosin X |
| chr11_+_57597563 | 0.70 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr18_+_44700796 | 0.70 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr14_+_35122660 | 0.69 |
ENST00000603544.5
|
PRORP
|
protein only RNase P catalytic subunit |
| chr17_-_59151794 | 0.69 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_+_155308930 | 0.67 |
ENST00000465559.5
ENST00000612683.1 |
FDPS
|
farnesyl diphosphate synthase |
| chr3_-_149377637 | 0.67 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr5_+_58491427 | 0.67 |
ENST00000396776.6
ENST00000502276.6 ENST00000511930.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr1_+_157993273 | 0.67 |
ENST00000360089.8
ENST00000368173.7 |
KIRREL1
|
kirre like nephrin family adhesion molecule 1 |
| chr17_-_445939 | 0.67 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr5_+_58491451 | 0.65 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr4_+_155666718 | 0.64 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr13_-_23433676 | 0.64 |
ENST00000682547.1
ENST00000455470.6 ENST00000382292.9 |
SACS
|
sacsin molecular chaperone |
| chr11_-_7796942 | 0.64 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2 |
| chr1_-_153150884 | 0.63 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr15_-_19988117 | 0.62 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr12_-_68225806 | 0.62 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr11_+_55827219 | 0.61 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor family 5 subfamily L member 2 |
| chr1_+_155308748 | 0.60 |
ENST00000611010.4
ENST00000447866.5 ENST00000368356.9 ENST00000467076.5 ENST00000491013.5 ENST00000356657.10 |
FDPS
|
farnesyl diphosphate synthase |
| chr11_-_14891643 | 0.60 |
ENST00000532378.5
|
CYP2R1
|
cytochrome P450 family 2 subfamily R member 1 |
| chr13_-_23433735 | 0.60 |
ENST00000423156.2
ENST00000683210.1 ENST00000682775.1 ENST00000684497.1 ENST00000682944.1 ENST00000683489.1 ENST00000684385.1 ENST00000683680.1 |
SACS
|
sacsin molecular chaperone |
| chr14_-_106211453 | 0.60 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr13_-_75366973 | 0.60 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chrX_+_7219431 | 0.59 |
ENST00000674499.1
ENST00000217961.5 |
STS
|
steroid sulfatase |
| chr4_-_103077282 | 0.59 |
ENST00000503230.5
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9 member B2 |
| chr18_-_26863187 | 0.58 |
ENST00000440832.7
|
AQP4
|
aquaporin 4 |
| chr17_-_47189176 | 0.58 |
ENST00000531206.5
ENST00000527547.5 ENST00000575483.5 ENST00000066544.8 |
CDC27
|
cell division cycle 27 |
| chr14_+_94561435 | 0.58 |
ENST00000557004.6
ENST00000555095.5 ENST00000298841.5 ENST00000554220.5 ENST00000553780.5 |
SERPINA4
SERPINA5
|
serpin family A member 4 serpin family A member 5 |
| chr2_-_40453438 | 0.57 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr8_-_7430348 | 0.57 |
ENST00000318124.3
|
DEFB103B
|
defensin beta 103B |
| chr2_-_89027700 | 0.56 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr19_-_42427379 | 0.56 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr3_-_20012250 | 0.56 |
ENST00000389050.5
|
PP2D1
|
protein phosphatase 2C like domain containing 1 |
| chr11_-_55936400 | 0.56 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr10_+_18400562 | 0.55 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr20_+_45416551 | 0.55 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chrX_-_103255117 | 0.55 |
ENST00000372685.8
ENST00000360000.8 ENST00000451678.1 |
TCEAL8
|
transcription elongation factor A like 8 |
| chr7_+_120988683 | 0.55 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr16_+_28878480 | 0.54 |
ENST00000395503.9
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr14_+_30577752 | 0.54 |
ENST00000547532.5
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr14_-_80231052 | 0.54 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr8_-_48921419 | 0.53 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr8_-_42768602 | 0.53 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr1_-_66801276 | 0.53 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr7_+_116222804 | 0.53 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr8_-_30812773 | 0.53 |
ENST00000221138.9
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr14_-_106593319 | 0.52 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr4_+_168631597 | 0.52 |
ENST00000504519.5
ENST00000512127.5 |
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_+_44844015 | 0.51 |
ENST00000339042.6
|
DUSP21
|
dual specificity phosphatase 21 |
| chr4_-_102828048 | 0.51 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr6_+_128883114 | 0.51 |
ENST00000421865.3
ENST00000618192.4 ENST00000617695.4 |
LAMA2
|
laminin subunit alpha 2 |
| chr8_-_33567118 | 0.50 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr4_+_118888918 | 0.50 |
ENST00000434046.6
|
SYNPO2
|
synaptopodin 2 |
| chr1_+_115029823 | 0.49 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr11_-_5301946 | 0.49 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr7_-_111392915 | 0.49 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr5_-_177462379 | 0.48 |
ENST00000512501.1
|
DBN1
|
drebrin 1 |
| chr17_+_47651061 | 0.48 |
ENST00000540627.5
|
KPNB1
|
karyopherin subunit beta 1 |
| chr8_+_104223320 | 0.48 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_+_43427535 | 0.48 |
ENST00000372530.9
|
ABCC10
|
ATP binding cassette subfamily C member 10 |
| chr19_-_3557563 | 0.47 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr1_-_201171545 | 0.47 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr4_-_102828022 | 0.47 |
ENST00000502690.5
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr3_+_111999326 | 0.46 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chrX_-_101617921 | 0.46 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr13_-_35476682 | 0.46 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chr15_-_91022540 | 0.46 |
ENST00000333371.8
ENST00000535906.1 |
VPS33B
|
VPS33B late endosome and lysosome associated |
| chr2_+_86907953 | 0.45 |
ENST00000409776.6
|
RGPD1
|
RANBP2 like and GRIP domain containing 1 |
| chr12_+_8157034 | 0.45 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr17_-_74776323 | 0.45 |
ENST00000582870.5
ENST00000581136.5 ENST00000579218.5 ENST00000583476.5 ENST00000580301.5 ENST00000583757.5 ENST00000357814.8 ENST00000582524.5 |
NAT9
|
N-acetyltransferase 9 (putative) |
| chr6_-_38703066 | 0.45 |
ENST00000373365.5
|
GLO1
|
glyoxalase I |
| chr15_+_94355956 | 0.45 |
ENST00000557742.1
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr16_-_55833085 | 0.44 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1 |
| chr8_+_7881387 | 0.44 |
ENST00000314357.4
|
DEFB103A
|
defensin beta 103A |
| chr4_+_70050431 | 0.44 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
| chr2_+_108377947 | 0.43 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr11_-_11353241 | 0.43 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr10_+_18260715 | 0.43 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr20_-_1466822 | 0.42 |
ENST00000353088.6
ENST00000216879.9 |
NSFL1C
|
NSFL1 cofactor |
| chr6_+_63521738 | 0.42 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr13_+_31739542 | 0.42 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr2_+_233729042 | 0.41 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr19_-_14848922 | 0.40 |
ENST00000641129.1
|
OR7A10
|
olfactory receptor family 7 subfamily A member 10 |
| chr12_-_11134644 | 0.40 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chrX_+_37780049 | 0.39 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr12_+_26011713 | 0.39 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr3_-_46208304 | 0.39 |
ENST00000296140.4
|
CCR1
|
C-C motif chemokine receptor 1 |
| chr1_+_76867469 | 0.38 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr3_+_111998739 | 0.38 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr17_+_41966787 | 0.38 |
ENST00000393892.8
ENST00000587679.1 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr4_+_89901979 | 0.38 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr4_-_89836213 | 0.37 |
ENST00000618500.4
ENST00000508895.5 |
SNCA
|
synuclein alpha |
| chr3_+_111999189 | 0.37 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr17_+_35587239 | 0.37 |
ENST00000621914.4
ENST00000621668.4 ENST00000616681.4 ENST00000612035.4 ENST00000610402.5 ENST00000614600.4 ENST00000590432.5 ENST00000612116.5 |
AP2B1
|
adaptor related protein complex 2 subunit beta 1 |
| chr2_+_90069662 | 0.37 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chrX_+_30243715 | 0.37 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr17_+_41966814 | 0.36 |
ENST00000393888.1
ENST00000441615.2 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr19_-_35812838 | 0.36 |
ENST00000653904.2
|
PRODH2
|
proline dehydrogenase 2 |
| chr20_-_35147285 | 0.36 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr7_+_141738321 | 0.36 |
ENST00000612337.4
|
SSBP1
|
single stranded DNA binding protein 1 |
| chr14_+_35122722 | 0.36 |
ENST00000605870.5
ENST00000557404.3 |
PRORP
|
protein only RNase P catalytic subunit |
| chrX_-_139642889 | 0.35 |
ENST00000370576.9
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr20_-_1466904 | 0.35 |
ENST00000476071.5
|
NSFL1C
|
NSFL1 cofactor |
| chr13_+_30422487 | 0.35 |
ENST00000638137.1
ENST00000635918.2 |
UBE2L5
|
ubiquitin conjugating enzyme E2 L5 |
| chr3_-_151316795 | 0.35 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr14_+_71933116 | 0.34 |
ENST00000553530.5
ENST00000556437.5 |
RGS6
|
regulator of G protein signaling 6 |
| chr19_-_43883964 | 0.34 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr4_+_109827963 | 0.34 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr12_-_91058016 | 0.34 |
ENST00000266719.4
|
KERA
|
keratocan |
| chrX_+_154304923 | 0.34 |
ENST00000426989.5
ENST00000426203.5 ENST00000369912.2 |
TKTL1
|
transketolase like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.7 | 7.9 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 3.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.5 | 1.9 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.5 | 2.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 1.6 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.3 | 5.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 1.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 1.3 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 1.9 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.2 | 0.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 0.7 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.2 | 1.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.2 | 5.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.2 | 0.7 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.2 | 0.5 | GO:0031443 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 0.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 1.8 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 2.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 2.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 1.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 2.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.4 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 1.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.6 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 2.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.5 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.6 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.7 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 1.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 3.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.2 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.1 | 0.5 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.6 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.2 | GO:0014034 | neural crest cell fate commitment(GO:0014034) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 1.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.4 | GO:2000470 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 1.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.8 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.1 | 0.4 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 1.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.0 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 2.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 5.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.5 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.7 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.3 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 1.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.2 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 3.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.6 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 2.2 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 2.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.5 | GO:0055026 | negative regulation of cardiac muscle tissue development(GO:0055026) |
| 0.0 | 0.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.9 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.8 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0002839 | positive regulation of response to tumor cell(GO:0002836) positive regulation of immune response to tumor cell(GO:0002839) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.5 | 8.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 1.2 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.3 | 0.9 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 1.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 0.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 0.8 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 2.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.6 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 1.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 3.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 2.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 3.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 6.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 1.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.7 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 1.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.1 | 5.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.6 | 8.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 1.4 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.3 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.3 | 1.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 0.7 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.2 | 2.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.6 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 3.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 0.6 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.6 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.1 | 0.7 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 2.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.9 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 2.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 2.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 1.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 1.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 7.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 2.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 2.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 2.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 4.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 6.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 8.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.8 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 2.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |