Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HOXC10_HOXD13
Z-value: 1.05
Transcription factors associated with HOXC10_HOXD13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
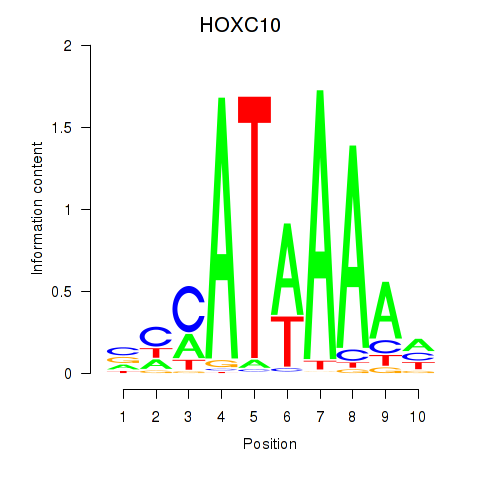
HOXC10
|
ENSG00000180818.5 | HOXC10 |
|
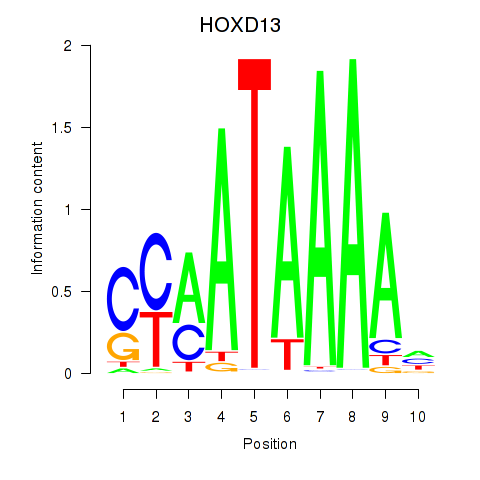
HOXD13
|
ENSG00000128714.6 | HOXD13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD13 | hg38_v1_chr2_+_176092715_176092729 | 0.72 | 8.6e-06 | Click! |
| HOXC10 | hg38_v1_chr12_+_53985138_53985183 | 0.08 | 6.7e-01 | Click! |
Activity profile of HOXC10_HOXD13 motif
Sorted Z-values of HOXC10_HOXD13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC10_HOXD13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_215436061 | 10.15 |
ENST00000421182.5
ENST00000432072.6 ENST00000323926.10 ENST00000336916.8 ENST00000357867.8 ENST00000359671.5 ENST00000446046.5 ENST00000354785.11 ENST00000356005.8 ENST00000443816.5 ENST00000426059.1 |
FN1
|
fibronectin 1 |
| chr11_+_111245725 | 6.47 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr12_-_52385649 | 5.92 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr2_-_189179754 | 5.89 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr12_-_91111460 | 4.75 |
ENST00000266718.5
|
LUM
|
lumican |
| chr1_+_84164962 | 3.59 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr7_-_76626127 | 2.75 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr1_+_152985231 | 2.53 |
ENST00000368762.1
|
SPRR1A
|
small proline rich protein 1A |
| chr7_-_41703062 | 2.36 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr7_+_134779625 | 2.33 |
ENST00000454108.5
ENST00000361675.7 |
CALD1
|
caldesmon 1 |
| chr1_-_152414256 | 2.32 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr13_-_20232191 | 2.21 |
ENST00000647243.1
|
GJB6
|
gap junction protein beta 6 |
| chr9_+_102995308 | 2.21 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr12_-_91146195 | 2.17 |
ENST00000548218.1
|
DCN
|
decorin |
| chr2_+_157257687 | 2.13 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr3_+_57890011 | 2.11 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr7_+_134779663 | 2.09 |
ENST00000361901.6
|
CALD1
|
caldesmon 1 |
| chr20_+_38926312 | 2.09 |
ENST00000619304.4
ENST00000619850.2 |
FAM83D
|
family with sequence similarity 83 member D |
| chr20_+_59604527 | 2.04 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr17_-_40984297 | 2.04 |
ENST00000377755.9
|
KRT40
|
keratin 40 |
| chr4_+_68447453 | 2.01 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr6_+_12290353 | 1.98 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chr2_-_216013582 | 1.96 |
ENST00000620139.4
|
MREG
|
melanoregulin |
| chr15_-_83283449 | 1.84 |
ENST00000569704.2
|
BNC1
|
basonuclin 1 |
| chr1_+_84164370 | 1.81 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr21_+_38272291 | 1.79 |
ENST00000438657.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr4_-_48016631 | 1.78 |
ENST00000513178.2
ENST00000514170.7 |
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr4_-_67883987 | 1.71 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr18_-_12656716 | 1.69 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr15_-_79971164 | 1.64 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr1_+_31576485 | 1.59 |
ENST00000457433.6
ENST00000271064.12 |
TINAGL1
|
tubulointerstitial nephritis antigen like 1 |
| chr12_-_95116967 | 1.55 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr3_-_52056552 | 1.53 |
ENST00000495880.2
|
DUSP7
|
dual specificity phosphatase 7 |
| chr4_+_85604146 | 1.53 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_-_108439472 | 1.51 |
ENST00000372216.8
|
COL4A6
|
collagen type IV alpha 6 chain |
| chr11_-_128522264 | 1.45 |
ENST00000531611.5
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr1_+_116111395 | 1.42 |
ENST00000684484.1
ENST00000369500.4 |
MAB21L3
|
mab-21 like 3 |
| chr17_-_41369807 | 1.42 |
ENST00000251646.8
|
KRT33B
|
keratin 33B |
| chr16_-_56425424 | 1.42 |
ENST00000290649.10
|
AMFR
|
autocrine motility factor receptor |
| chr1_+_209686173 | 1.39 |
ENST00000615289.4
ENST00000367028.6 ENST00000261465.5 |
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr1_+_153031195 | 1.39 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B |
| chr21_+_38272250 | 1.38 |
ENST00000398932.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr21_+_38272410 | 1.37 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr8_+_31639755 | 1.35 |
ENST00000520407.5
|
NRG1
|
neuregulin 1 |
| chr12_-_84892120 | 1.35 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr17_-_40703744 | 1.34 |
ENST00000264651.3
|
KRT24
|
keratin 24 |
| chr1_-_205449924 | 1.31 |
ENST00000367154.5
|
LEMD1
|
LEM domain containing 1 |
| chr20_+_3786772 | 1.29 |
ENST00000344256.10
ENST00000379598.9 |
CDC25B
|
cell division cycle 25B |
| chr11_-_128522285 | 1.28 |
ENST00000319397.6
ENST00000535549.5 |
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr11_-_107858777 | 1.26 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr20_+_63696643 | 1.26 |
ENST00000369996.3
|
TNFRSF6B
|
TNF receptor superfamily member 6b |
| chr4_-_68245683 | 1.26 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B |
| chr12_+_92702843 | 1.22 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr7_+_134866831 | 1.20 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr7_+_134891566 | 1.20 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr12_+_130162456 | 1.18 |
ENST00000539839.1
ENST00000229030.5 |
FZD10
|
frizzled class receptor 10 |
| chr3_-_185821092 | 1.18 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr18_-_5396265 | 1.17 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr4_-_39032343 | 1.17 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr2_-_207167220 | 1.15 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr2_+_209579399 | 1.15 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr20_+_44355692 | 1.12 |
ENST00000316673.8
ENST00000609795.5 ENST00000457232.5 ENST00000609262.5 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr21_-_26845402 | 1.12 |
ENST00000284984.8
ENST00000676955.1 |
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr3_+_101827982 | 1.11 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr12_-_30735014 | 1.11 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr2_-_216013517 | 1.10 |
ENST00000263268.11
|
MREG
|
melanoregulin |
| chr7_+_1688119 | 1.07 |
ENST00000424383.4
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr6_-_27146841 | 1.07 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr3_-_116444983 | 1.05 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr15_+_67125707 | 0.98 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr18_+_36544544 | 0.97 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr6_+_25726767 | 0.96 |
ENST00000274764.5
|
H2BC1
|
H2B clustered histone 1 |
| chr17_-_41397600 | 0.95 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr16_+_55566668 | 0.94 |
ENST00000457326.3
|
CAPNS2
|
calpain small subunit 2 |
| chr3_-_195583931 | 0.93 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr12_+_92702983 | 0.93 |
ENST00000344636.6
ENST00000544406.2 |
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr3_-_48016400 | 0.93 |
ENST00000434267.5
ENST00000683076.1 ENST00000633710.1 |
MAP4
|
microtubule associated protein 4 |
| chr4_+_48016764 | 0.92 |
ENST00000295461.10
|
NIPAL1
|
NIPA like domain containing 1 |
| chr2_+_9961165 | 0.91 |
ENST00000405379.6
|
GRHL1
|
grainyhead like transcription factor 1 |
| chr1_+_74235377 | 0.91 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr2_+_209579598 | 0.89 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr1_-_9069608 | 0.89 |
ENST00000377424.9
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr8_+_12108172 | 0.88 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D |
| chr12_-_53220229 | 0.87 |
ENST00000338561.9
|
RARG
|
retinoic acid receptor gamma |
| chr20_+_16748358 | 0.86 |
ENST00000246081.3
|
OTOR
|
otoraplin |
| chr21_-_26843012 | 0.85 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 0.84 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr2_+_33134620 | 0.84 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr11_+_63838902 | 0.84 |
ENST00000377810.8
|
MARK2
|
microtubule affinity regulating kinase 2 |
| chr1_-_28058087 | 0.84 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr17_-_41489907 | 0.83 |
ENST00000328119.11
|
KRT36
|
keratin 36 |
| chr6_-_27807916 | 0.82 |
ENST00000377401.3
|
H2BC13
|
H2B clustered histone 13 |
| chr15_+_67138001 | 0.82 |
ENST00000439724.7
|
SMAD3
|
SMAD family member 3 |
| chr9_+_122371036 | 0.82 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr11_-_11353241 | 0.82 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr7_+_134891400 | 0.81 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr1_-_155910881 | 0.81 |
ENST00000609492.1
ENST00000368322.7 |
RIT1
|
Ras like without CAAX 1 |
| chr22_+_22906342 | 0.80 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr15_+_41256907 | 0.79 |
ENST00000560965.1
|
CHP1
|
calcineurin like EF-hand protein 1 |
| chr1_+_50109817 | 0.77 |
ENST00000652353.1
ENST00000371821.6 ENST00000652274.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr19_+_47994696 | 0.77 |
ENST00000596043.5
ENST00000597519.5 |
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr18_+_63777773 | 0.76 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr19_+_47994625 | 0.76 |
ENST00000339841.7
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr15_-_74725370 | 0.76 |
ENST00000567032.5
ENST00000564596.5 ENST00000566503.1 ENST00000395049.8 ENST00000379727.8 ENST00000617691.4 ENST00000395048.6 |
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1 |
| chr12_-_8662073 | 0.76 |
ENST00000535411.5
ENST00000540087.5 |
MFAP5
|
microfibril associated protein 5 |
| chr2_+_33134579 | 0.75 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_91182784 | 0.74 |
ENST00000547568.6
ENST00000052754.10 ENST00000552962.5 |
DCN
|
decorin |
| chr2_+_100974849 | 0.73 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr1_-_93681829 | 0.73 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr17_-_41350824 | 0.73 |
ENST00000007735.4
|
KRT33A
|
keratin 33A |
| chr19_+_926001 | 0.73 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr3_+_160677152 | 0.72 |
ENST00000320767.4
|
ARL14
|
ADP ribosylation factor like GTPase 14 |
| chr21_-_30487436 | 0.71 |
ENST00000334055.5
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr11_-_128522189 | 0.71 |
ENST00000526145.6
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr1_-_9069572 | 0.70 |
ENST00000377414.7
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr11_-_5516690 | 0.69 |
ENST00000380184.2
|
UBQLNL
|
ubiquilin like |
| chr17_-_41481140 | 0.69 |
ENST00000246639.6
ENST00000393989.1 |
KRT35
|
keratin 35 |
| chr4_-_47981535 | 0.68 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr9_+_128322540 | 0.67 |
ENST00000609948.1
ENST00000608951.5 |
COQ4
|
coenzyme Q4 |
| chr2_-_237414157 | 0.67 |
ENST00000295550.9
ENST00000353578.9 ENST00000392004.7 ENST00000433762.1 ENST00000392003.6 |
COL6A3
|
collagen type VI alpha 3 chain |
| chr22_+_50170720 | 0.66 |
ENST00000159647.9
ENST00000395842.3 |
PANX2
|
pannexin 2 |
| chr1_-_158554405 | 0.66 |
ENST00000641282.1
ENST00000641622.1 |
OR6Y1
|
olfactory receptor family 6 subfamily Y member 1 |
| chr2_-_29223808 | 0.65 |
ENST00000642122.1
|
ALK
|
ALK receptor tyrosine kinase |
| chr5_-_141673160 | 0.65 |
ENST00000513878.5
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr20_-_57710539 | 0.64 |
ENST00000395816.7
ENST00000347215.8 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_-_111567271 | 0.63 |
ENST00000464903.1
ENST00000368730.5 ENST00000368735.1 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr7_+_80646436 | 0.62 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr6_+_27815010 | 0.62 |
ENST00000621112.2
|
H2BC14
|
H2B clustered histone 14 |
| chr5_-_88785493 | 0.62 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr10_-_29634964 | 0.60 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr12_-_52321395 | 0.59 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr17_+_17179527 | 0.59 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr11_-_55936400 | 0.56 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr17_+_59940908 | 0.56 |
ENST00000591035.1
|
ENSG00000267318.1
|
novel protein |
| chr6_-_111567120 | 0.56 |
ENST00000368734.5
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr19_+_35143237 | 0.56 |
ENST00000586063.5
ENST00000270310.7 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr8_-_59119121 | 0.56 |
ENST00000361421.2
|
TOX
|
thymocyte selection associated high mobility group box |
| chr10_+_88594746 | 0.55 |
ENST00000531458.1
|
LIPJ
|
lipase family member J |
| chr14_+_21057822 | 0.55 |
ENST00000308227.2
|
RNASE8
|
ribonuclease A family member 8 |
| chr15_-_55249029 | 0.54 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_+_54100161 | 0.54 |
ENST00000326902.7
ENST00000503800.1 |
GSX2
|
GS homeobox 2 |
| chr2_-_70553440 | 0.54 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr2_+_182716227 | 0.54 |
ENST00000680258.1
ENST00000680667.1 ENST00000264065.12 ENST00000616986.5 ENST00000679884.1 |
DNAJC10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr2_-_55334529 | 0.54 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr2_+_29113989 | 0.54 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr7_-_27165517 | 0.53 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr14_-_50561119 | 0.53 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_+_84181630 | 0.53 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr8_+_12104389 | 0.52 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr1_+_50105666 | 0.52 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr6_+_130018565 | 0.52 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr1_+_192158448 | 0.52 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr11_+_64234569 | 0.52 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr7_+_80646305 | 0.51 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr5_+_31193678 | 0.51 |
ENST00000265071.3
|
CDH6
|
cadherin 6 |
| chr12_+_112938284 | 0.51 |
ENST00000681346.1
|
OAS3
|
2'-5'-oligoadenylate synthetase 3 |
| chr12_-_91179472 | 0.51 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr14_+_51847145 | 0.50 |
ENST00000615906.4
|
GNG2
|
G protein subunit gamma 2 |
| chr13_+_108596152 | 0.50 |
ENST00000356711.7
ENST00000251041.10 |
MYO16
|
myosin XVI |
| chr8_+_76681208 | 0.49 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr2_+_209579429 | 0.49 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2 |
| chr20_-_52105644 | 0.49 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr19_-_10309783 | 0.49 |
ENST00000403352.1
ENST00000403903.7 |
ZGLP1
|
zinc finger GATA like protein 1 |
| chr1_-_43453792 | 0.48 |
ENST00000372434.5
ENST00000486909.1 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr12_-_18090141 | 0.47 |
ENST00000536890.1
|
RERGL
|
RERG like |
| chrX_+_66164210 | 0.47 |
ENST00000343002.7
ENST00000336279.9 |
HEPH
|
hephaestin |
| chr7_-_48029102 | 0.47 |
ENST00000297325.9
ENST00000412142.5 ENST00000395572.6 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr9_+_122371014 | 0.46 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr12_-_27971970 | 0.46 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr10_-_29736956 | 0.46 |
ENST00000674475.1
|
SVIL
|
supervillin |
| chr4_+_70383123 | 0.46 |
ENST00000304915.8
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr7_+_16661182 | 0.46 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr3_+_189789643 | 0.46 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chr3_-_123620571 | 0.45 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr3_-_179243284 | 0.45 |
ENST00000486944.2
|
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr3_-_33645433 | 0.45 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_-_173399102 | 0.45 |
ENST00000296506.8
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr12_+_71439976 | 0.45 |
ENST00000536515.5
ENST00000540815.2 |
LGR5
|
leucine rich repeat containing G protein-coupled receptor 5 |
| chr5_+_96662046 | 0.45 |
ENST00000338252.7
ENST00000508830.5 |
CAST
|
calpastatin |
| chr14_-_106715166 | 0.44 |
ENST00000390633.2
|
IGHV1-69
|
immunoglobulin heavy variable 1-69 |
| chr4_+_87799546 | 0.44 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr8_+_124474843 | 0.44 |
ENST00000303545.4
|
RNF139
|
ring finger protein 139 |
| chr2_+_172928165 | 0.44 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chrX_+_47585212 | 0.44 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr9_+_122370523 | 0.44 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr10_-_13972355 | 0.44 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr14_-_106762576 | 0.44 |
ENST00000624687.1
|
IGHV1-69D
|
immunoglobulin heavy variable 1-69D |
| chr3_-_58577648 | 0.43 |
ENST00000394481.5
|
FAM107A
|
family with sequence similarity 107 member A |
| chr3_-_123620496 | 0.43 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr1_-_209784521 | 0.43 |
ENST00000294811.2
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr9_-_111759508 | 0.43 |
ENST00000394777.8
ENST00000394779.7 |
SHOC1
|
shortage in chiasmata 1 |
| chr18_+_63887698 | 0.43 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr2_-_65366650 | 0.43 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chrX_+_66164340 | 0.43 |
ENST00000441993.7
ENST00000419594.6 ENST00000425114.2 |
HEPH
|
hephaestin |
| chr17_-_64263221 | 0.42 |
ENST00000258991.7
ENST00000583738.1 ENST00000584379.6 |
TEX2
|
testis expressed 2 |
| chr9_-_92878018 | 0.42 |
ENST00000332591.6
ENST00000375495.8 ENST00000395505.6 ENST00000395506.7 |
ZNF484
|
zinc finger protein 484 |
| chr2_+_200440649 | 0.42 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chrM_-_14669 | 0.42 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 |
| chrX_+_136305988 | 0.42 |
ENST00000394141.1
|
ADGRG4
|
adhesion G protein-coupled receptor G4 |
| chr12_-_68225806 | 0.42 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 2.0 | 5.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 | 5.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.8 | 2.4 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.7 | 2.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.5 | 2.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 5.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.5 | 2.7 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.4 | 5.9 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 3.4 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 0.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 1.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 1.8 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.3 | 0.8 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.2 | 1.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 3.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.4 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.2 | 1.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 0.9 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 0.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 1.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 0.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.1 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 1.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.4 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 2.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.3 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.8 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 1.4 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 1.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.9 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.3 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.4 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 12.8 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.4 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) |
| 0.1 | 1.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 3.1 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.3 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.4 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.1 | 0.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 1.9 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 1.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.2 | GO:1901877 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 4.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.1 | GO:0045608 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.5 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 0.2 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 1.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0002035 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 1.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.1 | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) |
| 0.1 | 0.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.4 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0061394 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.7 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.2 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.9 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.7 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.5 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 1.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:1900450 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) |
| 0.0 | 1.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 2.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.2 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 2.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.2 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.9 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.7 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.4 | GO:0021516 | grooming behavior(GO:0007625) dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.4 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 1.0 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.3 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.6 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 2.1 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.6 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.9 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 1.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.3 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.0 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0010888 | negative regulation of lipid storage(GO:0010888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.9 | 10.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 2.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.7 | 2.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.7 | 2.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 7.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 4.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 2.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 4.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 5.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 1.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 2.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.9 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.3 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 2.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 2.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 13.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.3 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 2.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 1.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 5.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 2.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 1.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 1.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.3 | 1.8 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.3 | 2.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 2.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 7.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 4.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.2 | 2.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 1.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.6 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 0.5 | GO:0008903 | hydroxypyruvate isomerase activity(GO:0008903) |
| 0.2 | 0.8 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 10.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 14.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 2.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 2.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 2.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 4.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 4.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 3.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.7 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 3.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 9.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 8.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 4.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 9.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 4.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 6.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 8.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 5.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 7.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.8 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 1.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |