Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for HOXC9
Z-value: 0.41
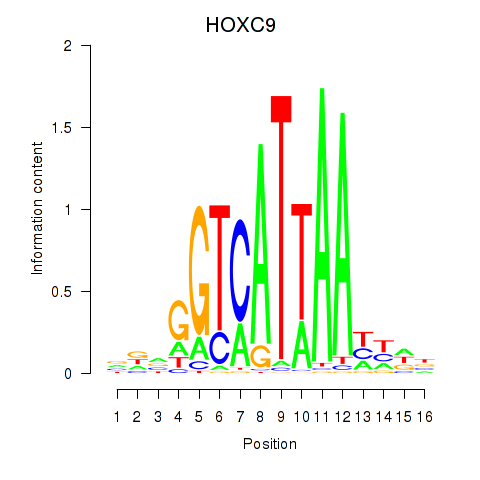
Transcription factors associated with HOXC9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC9
|
ENSG00000180806.5 | HOXC9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC9 | hg38_v1_chr12_+_54000096_54000180 | -0.31 | 9.2e-02 | Click! |
Activity profile of HOXC9 motif
Sorted Z-values of HOXC9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_55166632 | 1.20 |
ENST00000532817.5
ENST00000527223.6 ENST00000391720.8 |
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr19_-_55166671 | 1.19 |
ENST00000455045.5
|
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr19_-_55166565 | 1.17 |
ENST00000526003.5
ENST00000534170.5 ENST00000524407.7 |
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr6_+_52423680 | 0.85 |
ENST00000538167.2
|
EFHC1
|
EF-hand domain containing 1 |
| chr11_-_26572254 | 0.76 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr11_+_72189659 | 0.73 |
ENST00000393681.6
|
FOLR1
|
folate receptor alpha |
| chr11_+_72189528 | 0.72 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha |
| chr11_-_26572130 | 0.65 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_+_68815991 | 0.62 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr11_-_26572102 | 0.57 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr13_+_35476740 | 0.53 |
ENST00000537702.5
|
NBEA
|
neurobeachin |
| chr17_+_70075215 | 0.51 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr14_-_106470788 | 0.50 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr2_-_159798043 | 0.50 |
ENST00000664982.1
ENST00000259053.6 |
ENSG00000287091.1
CD302
|
novel transcript, sense intronic to CD302and LY75-CD302 CD302 molecule |
| chr2_-_159798234 | 0.49 |
ENST00000429078.6
ENST00000553424.5 |
CD302
|
CD302 molecule |
| chrX_+_153072454 | 0.49 |
ENST00000421798.5
|
PNMA6A
|
PNMA family member 6A |
| chr3_-_45884685 | 0.43 |
ENST00000684620.1
|
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr6_+_32844789 | 0.42 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr22_+_23145366 | 0.41 |
ENST00000341989.9
ENST00000263116.8 |
RAB36
|
RAB36, member RAS oncogene family |
| chr3_-_112829367 | 0.38 |
ENST00000448932.4
ENST00000617549.3 |
CD200R1L
|
CD200 receptor 1 like |
| chr7_-_130441136 | 0.35 |
ENST00000675596.1
ENST00000676312.1 |
CEP41
|
centrosomal protein 41 |
| chr12_+_20810698 | 0.34 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr12_-_62935117 | 0.34 |
ENST00000228705.7
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent 1H |
| chr17_+_70075317 | 0.32 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr7_+_150685693 | 0.31 |
ENST00000223293.10
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr1_+_63523490 | 0.30 |
ENST00000371088.5
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr3_-_108953870 | 0.29 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr16_+_28494634 | 0.28 |
ENST00000564831.6
ENST00000431282.2 |
APOBR
|
apolipoprotein B receptor |
| chr5_+_157269317 | 0.26 |
ENST00000618329.4
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr16_-_56519943 | 0.26 |
ENST00000683875.1
|
BBS2
|
Bardet-Biedl syndrome 2 |
| chr16_-_56520087 | 0.26 |
ENST00000682737.1
|
BBS2
|
Bardet-Biedl syndrome 2 |
| chr1_+_40396766 | 0.26 |
ENST00000539317.2
|
SMAP2
|
small ArfGAP2 |
| chr22_-_37844308 | 0.25 |
ENST00000411961.6
ENST00000434930.1 ENST00000215941.9 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr8_-_71361860 | 0.25 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_+_210328244 | 0.25 |
ENST00000541565.5
ENST00000413764.6 |
HHAT
|
hedgehog acyltransferase |
| chr19_-_14884761 | 0.24 |
ENST00000642123.1
|
OR7A17
|
olfactory receptor family 7 subfamily A member 17 |
| chr22_+_31754862 | 0.24 |
ENST00000382111.6
ENST00000645407.1 ENST00000646701.1 |
DEPDC5
ENSG00000285404.1
|
DEP domain containing 5, GATOR1 subcomplex subunit novel protein, DEPDC5-YWHAH readthrough |
| chr1_+_108560031 | 0.24 |
ENST00000405454.1
ENST00000370035.8 |
FAM102B
|
family with sequence similarity 102 member B |
| chr3_-_108953762 | 0.24 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr6_+_83193331 | 0.24 |
ENST00000369724.5
|
RWDD2A
|
RWD domain containing 2A |
| chr12_+_25052634 | 0.23 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr6_-_52994248 | 0.23 |
ENST00000457564.1
ENST00000370960.5 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr10_+_125896549 | 0.23 |
ENST00000368693.6
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr22_+_31753867 | 0.23 |
ENST00000535622.6
ENST00000645693.1 ENST00000642974.1 ENST00000645711.1 ENST00000644331.1 ENST00000645560.1 ENST00000437411.6 ENST00000433147.2 ENST00000646755.1 ENST00000382112.8 ENST00000647438.1 ENST00000400248.7 ENST00000456178.6 ENST00000646969.1 ENST00000642696.1 ENST00000647343.1 ENST00000400242.8 ENST00000651528.2 ENST00000645015.1 ENST00000645564.1 ENST00000646465.1 ENST00000400249.7 |
DEPDC5
|
DEP domain containing 5, GATOR1 subcomplex subunit |
| chr6_-_111483700 | 0.22 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr7_+_114416286 | 0.22 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2 |
| chr14_+_30577752 | 0.21 |
ENST00000547532.5
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr8_+_75539862 | 0.21 |
ENST00000396423.4
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr3_+_56557155 | 0.21 |
ENST00000422222.5
ENST00000394672.8 ENST00000326595.11 |
CCDC66
|
coiled-coil domain containing 66 |
| chr19_+_49513353 | 0.19 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr5_-_135954962 | 0.19 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chrX_+_66164340 | 0.19 |
ENST00000441993.7
ENST00000419594.6 ENST00000425114.2 |
HEPH
|
hephaestin |
| chr8_+_75539893 | 0.18 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr5_+_98773651 | 0.18 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr5_+_36606355 | 0.18 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chrX_+_66164210 | 0.17 |
ENST00000343002.7
ENST00000336279.9 |
HEPH
|
hephaestin |
| chr3_+_138621225 | 0.16 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr7_-_44082464 | 0.16 |
ENST00000335195.10
ENST00000395831.7 ENST00000414235.5 ENST00000242248.10 ENST00000452049.1 |
POLM
|
DNA polymerase mu |
| chrX_+_66162663 | 0.16 |
ENST00000519389.6
|
HEPH
|
hephaestin |
| chr8_-_92966129 | 0.16 |
ENST00000522925.5
ENST00000522903.5 ENST00000537541.1 ENST00000521988.6 ENST00000518748.5 ENST00000519069.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_+_25052512 | 0.16 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr3_+_185328886 | 0.16 |
ENST00000428617.1
ENST00000443863.5 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr10_+_76318330 | 0.16 |
ENST00000496424.2
|
LRMDA
|
leucine rich melanocyte differentiation associated |
| chr17_+_69502397 | 0.15 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr10_-_113854368 | 0.15 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr2_+_108378176 | 0.15 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr10_-_13302341 | 0.15 |
ENST00000396920.7
|
PHYH
|
phytanoyl-CoA 2-hydroxylase |
| chr11_-_129024157 | 0.15 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_+_138621207 | 0.15 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_160400543 | 0.15 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr8_-_17697654 | 0.14 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr3_+_41200080 | 0.14 |
ENST00000644524.1
|
CTNNB1
|
catenin beta 1 |
| chr14_-_21024092 | 0.14 |
ENST00000554398.5
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21023954 | 0.14 |
ENST00000554094.5
|
NDRG2
|
NDRG family member 2 |
| chr8_-_92966081 | 0.14 |
ENST00000517858.5
ENST00000378861.9 |
TRIQK
|
triple QxxK/R motif containing |
| chr11_+_7597182 | 0.14 |
ENST00000528883.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr8_-_92966105 | 0.14 |
ENST00000524037.5
ENST00000520430.5 ENST00000521617.5 ENST00000523580.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr1_-_158686700 | 0.14 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1 |
| chr9_-_20382461 | 0.14 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr11_+_73787853 | 0.14 |
ENST00000310614.12
ENST00000497094.6 ENST00000411840.6 ENST00000535277.5 ENST00000398483.7 ENST00000542303.5 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr11_-_5154757 | 0.13 |
ENST00000380367.3
|
OR52A1
|
olfactory receptor family 52 subfamily A member 1 |
| chr18_+_34978244 | 0.13 |
ENST00000436190.6
|
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr1_+_232950580 | 0.13 |
ENST00000366628.10
ENST00000366627.4 |
NTPCR
|
nucleoside-triphosphatase, cancer-related |
| chr3_+_41200104 | 0.13 |
ENST00000643297.1
ENST00000450969.6 |
CTNNB1
|
catenin beta 1 |
| chr8_-_17722217 | 0.13 |
ENST00000381861.7
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr2_+_108377947 | 0.13 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr10_+_4786624 | 0.13 |
ENST00000533295.5
|
AKR1E2
|
aldo-keto reductase family 1 member E2 |
| chr6_-_116060859 | 0.13 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr1_+_158289916 | 0.12 |
ENST00000368170.8
|
CD1C
|
CD1c molecule |
| chr9_-_15472732 | 0.12 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr10_-_112183698 | 0.12 |
ENST00000369425.5
ENST00000348367.9 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr9_-_135961310 | 0.12 |
ENST00000371756.4
|
UBAC1
|
UBA domain containing 1 |
| chr8_+_106726012 | 0.12 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr4_-_69214743 | 0.12 |
ENST00000446444.2
|
UGT2B11
|
UDP glucuronosyltransferase family 2 member B11 |
| chr19_-_48249841 | 0.12 |
ENST00000447740.6
|
CARD8
|
caspase recruitment domain family member 8 |
| chr20_+_2207217 | 0.12 |
ENST00000447956.5
ENST00000411839.1 ENST00000651531.1 |
ENSG00000226644.5
ENSG00000286022.1
|
novel transcript novel protein |
| chr1_+_220690354 | 0.12 |
ENST00000294889.6
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr16_-_56519971 | 0.12 |
ENST00000682482.1
ENST00000682047.1 ENST00000682205.1 ENST00000682855.1 ENST00000245157.11 ENST00000682470.1 ENST00000682360.1 ENST00000569941.6 ENST00000568104.6 ENST00000684635.1 ENST00000682188.1 ENST00000683858.1 |
BBS2
|
Bardet-Biedl syndrome 2 |
| chr10_-_27240505 | 0.12 |
ENST00000375888.5
ENST00000676732.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr6_+_31655888 | 0.12 |
ENST00000375916.4
|
APOM
|
apolipoprotein M |
| chr18_+_58341038 | 0.12 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr10_+_113854610 | 0.11 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr1_-_53889766 | 0.11 |
ENST00000371399.5
ENST00000072644.7 |
YIPF1
|
Yip1 domain family member 1 |
| chr12_+_19205294 | 0.11 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr8_+_106726115 | 0.11 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr12_-_15221394 | 0.11 |
ENST00000537647.5
ENST00000256953.6 ENST00000546331.5 |
RERG
|
RAS like estrogen regulated growth inhibitor |
| chr15_-_44711306 | 0.11 |
ENST00000682850.1
|
PATL2
|
PAT1 homolog 2 |
| chr11_-_63144221 | 0.11 |
ENST00000417740.5
ENST00000612278.4 ENST00000326192.5 |
SLC22A24
|
solute carrier family 22 member 24 |
| chr14_+_51860632 | 0.11 |
ENST00000555472.5
ENST00000556766.5 |
GNG2
|
G protein subunit gamma 2 |
| chr11_-_119030848 | 0.10 |
ENST00000330775.9
ENST00000357590.9 ENST00000538950.5 ENST00000545985.5 |
SLC37A4
|
solute carrier family 37 member 4 |
| chr7_-_38265678 | 0.10 |
ENST00000443402.6
|
TRGC1
|
T cell receptor gamma constant 1 |
| chr13_+_105466216 | 0.10 |
ENST00000375936.8
ENST00000329625.9 ENST00000595812.2 ENST00000618629.1 |
DAOA
|
D-amino acid oxidase activator |
| chr1_+_22636577 | 0.10 |
ENST00000374642.8
ENST00000438241.1 |
C1QA
|
complement C1q A chain |
| chrX_-_13817027 | 0.10 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr5_+_80654669 | 0.10 |
ENST00000667069.1
|
MSH3
|
mutS homolog 3 |
| chr2_-_171894227 | 0.10 |
ENST00000422440.7
|
SLC25A12
|
solute carrier family 25 member 12 |
| chr7_-_113919000 | 0.10 |
ENST00000284601.4
|
PPP1R3A
|
protein phosphatase 1 regulatory subunit 3A |
| chr19_+_21082224 | 0.10 |
ENST00000620627.1
|
ZNF714
|
zinc finger protein 714 |
| chr17_+_7407838 | 0.10 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr2_+_37950432 | 0.10 |
ENST00000407257.5
ENST00000417700.6 ENST00000234195.7 ENST00000442857.5 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr19_+_20923222 | 0.09 |
ENST00000597314.5
ENST00000328178.13 ENST00000601924.5 |
ZNF85
|
zinc finger protein 85 |
| chr7_-_144410227 | 0.09 |
ENST00000467773.1
ENST00000483238.5 |
NOBOX
|
NOBOX oogenesis homeobox |
| chr12_-_89656051 | 0.09 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr13_-_52450590 | 0.09 |
ENST00000378060.9
|
VPS36
|
vacuolar protein sorting 36 homolog |
| chr5_-_146182475 | 0.09 |
ENST00000674158.1
ENST00000674191.1 ENST00000274562.13 |
LARS1
|
leucyl-tRNA synthetase 1 |
| chr8_+_17027230 | 0.09 |
ENST00000318063.10
|
MICU3
|
mitochondrial calcium uptake family member 3 |
| chr3_+_113532508 | 0.09 |
ENST00000264852.9
|
SIDT1
|
SID1 transmembrane family member 1 |
| chr19_-_43935234 | 0.09 |
ENST00000269973.10
|
ZNF45
|
zinc finger protein 45 |
| chr12_-_89656093 | 0.09 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr13_+_48976597 | 0.09 |
ENST00000541916.5
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr11_-_114595750 | 0.09 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr16_-_50681328 | 0.09 |
ENST00000300590.7
|
SNX20
|
sorting nexin 20 |
| chr19_-_48255863 | 0.08 |
ENST00000522889.5
ENST00000519940.6 ENST00000520753.5 ENST00000519332.5 ENST00000521437.1 ENST00000520007.5 ENST00000651546.1 ENST00000521613.5 |
CARD8
|
caspase recruitment domain family member 8 |
| chr5_-_146182591 | 0.08 |
ENST00000510191.5
ENST00000674277.1 ENST00000674447.1 ENST00000674270.1 ENST00000394434.7 ENST00000674290.1 ENST00000674398.1 ENST00000674174.1 |
LARS1
|
leucyl-tRNA synthetase 1 |
| chr5_-_58999885 | 0.08 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D |
| chr11_+_560956 | 0.08 |
ENST00000397582.7
ENST00000397583.8 |
RASSF7
|
Ras association domain family member 7 |
| chr1_-_212791762 | 0.08 |
ENST00000626725.1
ENST00000366977.8 ENST00000366976.3 |
NSL1
|
NSL1 component of MIS12 kinetochore complex |
| chr16_+_68085861 | 0.08 |
ENST00000570212.5
ENST00000562926.5 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr1_+_26472405 | 0.08 |
ENST00000361427.6
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr11_-_6006946 | 0.08 |
ENST00000641156.1
ENST00000641835.1 |
OR56A4
|
olfactory receptor family 56 subfamily A member 4 |
| chr18_-_21703688 | 0.08 |
ENST00000584464.1
ENST00000578270.5 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr16_-_12803785 | 0.08 |
ENST00000433677.6
ENST00000261660.4 ENST00000381774.9 |
CPPED1
|
calcineurin like phosphoesterase domain containing 1 |
| chr11_-_4242640 | 0.08 |
ENST00000640805.1
|
SSU72P2
|
SSU72 pseudogene 2 |
| chr1_+_26472459 | 0.08 |
ENST00000619352.4
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr7_+_1044542 | 0.08 |
ENST00000444847.2
|
GPR146
|
G protein-coupled receptor 146 |
| chr11_-_114595777 | 0.07 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr9_-_14300231 | 0.07 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr1_+_248838211 | 0.07 |
ENST00000306562.8
|
ZNF672
|
zinc finger protein 672 |
| chr12_+_25195230 | 0.07 |
ENST00000381356.9
|
ETFRF1
|
electron transfer flavoprotein regulatory factor 1 |
| chr12_+_25195205 | 0.07 |
ENST00000557540.7
|
ETFRF1
|
electron transfer flavoprotein regulatory factor 1 |
| chr17_+_57085092 | 0.07 |
ENST00000575322.1
ENST00000337714.8 |
AKAP1
|
A-kinase anchoring protein 1 |
| chr10_+_24466487 | 0.07 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr8_+_131904071 | 0.07 |
ENST00000254624.10
ENST00000522709.5 |
EFR3A
|
EFR3 homolog A |
| chrX_-_135799001 | 0.07 |
ENST00000612878.4
|
CT45A6
|
cancer/testis antigen family 45 member A6 |
| chr1_+_212791828 | 0.07 |
ENST00000532324.5
ENST00000530441.5 ENST00000526641.5 ENST00000531963.5 ENST00000366973.8 ENST00000366974.9 ENST00000526997.5 ENST00000488246.6 |
TATDN3
|
TatD DNase domain containing 3 |
| chr20_+_6007245 | 0.07 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr13_+_21140572 | 0.07 |
ENST00000607003.5
ENST00000492245.5 |
SAP18
|
Sin3A associated protein 18 |
| chr10_-_125816510 | 0.07 |
ENST00000650587.1
|
UROS
|
uroporphyrinogen III synthase |
| chr11_+_60280577 | 0.07 |
ENST00000679988.1
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr8_-_18684033 | 0.07 |
ENST00000614430.3
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr22_-_32255344 | 0.07 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chr18_+_58196736 | 0.07 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr10_-_72088533 | 0.07 |
ENST00000373109.7
|
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr17_-_41168219 | 0.07 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr6_-_109381739 | 0.07 |
ENST00000504373.2
|
CD164
|
CD164 molecule |
| chr11_+_60280658 | 0.06 |
ENST00000337908.5
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr17_+_50746614 | 0.06 |
ENST00000513969.5
ENST00000503728.1 |
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr16_-_66730216 | 0.06 |
ENST00000569320.5
|
DYNC1LI2
|
dynein cytoplasmic 1 light intermediate chain 2 |
| chr12_+_25195252 | 0.06 |
ENST00000555711.6
ENST00000554266.6 ENST00000556351.6 ENST00000556927.6 ENST00000556402.6 ENST00000553788.6 |
ETFRF1
|
electron transfer flavoprotein regulatory factor 1 |
| chr2_+_218607861 | 0.06 |
ENST00000450993.7
|
PLCD4
|
phospholipase C delta 4 |
| chr10_-_125816596 | 0.06 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr8_-_18684093 | 0.06 |
ENST00000428502.6
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_+_101185029 | 0.06 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr14_+_55027576 | 0.06 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chrX_-_135833541 | 0.06 |
ENST00000620885.1
|
CT45A7
|
cancer/testis antigen family 45 member A7 |
| chr5_-_148654522 | 0.06 |
ENST00000377888.8
|
HTR4
|
5-hydroxytryptamine receptor 4 |
| chr1_-_92486916 | 0.06 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr5_+_62578810 | 0.06 |
ENST00000334994.6
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr5_-_148654130 | 0.06 |
ENST00000360693.7
|
HTR4
|
5-hydroxytryptamine receptor 4 |
| chr11_+_22666604 | 0.05 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr4_-_68349750 | 0.05 |
ENST00000579690.5
|
YTHDC1
|
YTH domain containing 1 |
| chr17_-_29930062 | 0.05 |
ENST00000579954.1
ENST00000269033.7 ENST00000540801.6 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr2_+_232526153 | 0.05 |
ENST00000449596.5
ENST00000258385.8 ENST00000543200.5 |
CHRND
|
cholinergic receptor nicotinic delta subunit |
| chr2_+_108588453 | 0.05 |
ENST00000393310.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr1_+_12464912 | 0.05 |
ENST00000543766.2
|
VPS13D
|
vacuolar protein sorting 13 homolog D |
| chr17_+_44187210 | 0.05 |
ENST00000589785.1
ENST00000592825.1 ENST00000589184.5 |
TMUB2
|
transmembrane and ubiquitin like domain containing 2 |
| chr19_-_45424364 | 0.05 |
ENST00000589165.5
|
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr7_+_65373839 | 0.05 |
ENST00000431504.1
ENST00000328747.12 |
ZNF92
|
zinc finger protein 92 |
| chr15_+_67521104 | 0.05 |
ENST00000342683.6
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr14_+_73616844 | 0.05 |
ENST00000381139.1
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr14_+_20891385 | 0.05 |
ENST00000304639.4
|
RNASE3
|
ribonuclease A family member 3 |
| chr15_+_44711487 | 0.05 |
ENST00000544417.5
ENST00000559916.1 ENST00000648006.3 |
B2M
|
beta-2-microglobulin |
| chr2_+_225399684 | 0.05 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr5_-_35991433 | 0.05 |
ENST00000507113.5
ENST00000333811.5 |
UGT3A1
|
UDP glycosyltransferase family 3 member A1 |
| chr11_-_18526885 | 0.05 |
ENST00000251968.4
ENST00000536719.5 |
TSG101
|
tumor susceptibility 101 |
| chr4_-_117085541 | 0.05 |
ENST00000310754.5
|
TRAM1L1
|
translocation associated membrane protein 1 like 1 |
| chr2_+_201183120 | 0.04 |
ENST00000272879.9
ENST00000286186.11 ENST00000374650.7 ENST00000346817.9 ENST00000313728.11 ENST00000448480.1 |
CASP10
|
caspase 10 |
| chr6_+_29301701 | 0.04 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr2_-_208146150 | 0.04 |
ENST00000260988.5
|
CRYGB
|
crystallin gamma B |
| chr2_-_70248598 | 0.04 |
ENST00000445587.5
ENST00000433529.7 ENST00000415783.6 |
TIA1
|
TIA1 cytotoxic granule associated RNA binding protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.4 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 3.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.6 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.2 | GO:0021758 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.2 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.3 | GO:1904793 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:1904438 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 1.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:0006212 | uracil catabolic process(GO:0006212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |