Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
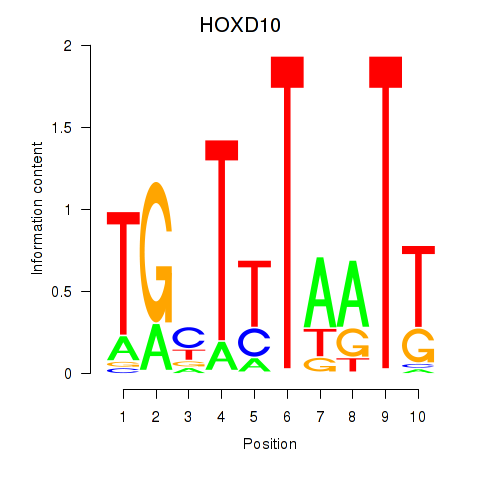
Results for HOXD10
Z-value: 0.64
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.6 | HOXD10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg38_v1_chr2_+_176116768_176116794 | -0.09 | 6.4e-01 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_47981535 | 1.03 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr4_-_67883987 | 0.88 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr5_+_96875978 | 0.75 |
ENST00000510373.5
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr18_+_63777773 | 0.74 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr15_-_28174423 | 0.65 |
ENST00000569772.1
|
HERC2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr12_-_13095664 | 0.64 |
ENST00000337630.10
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr18_+_31447732 | 0.60 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr1_-_94925759 | 0.59 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr13_+_77535742 | 0.57 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr13_+_77535681 | 0.57 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535669 | 0.57 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr4_+_186227501 | 0.52 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr2_-_29074515 | 0.49 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr5_-_135954962 | 0.49 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr10_-_77140757 | 0.48 |
ENST00000637862.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr10_-_89207306 | 0.47 |
ENST00000371852.4
|
CH25H
|
cholesterol 25-hydroxylase |
| chr2_-_29223808 | 0.47 |
ENST00000642122.1
|
ALK
|
ALK receptor tyrosine kinase |
| chr4_-_76007501 | 0.46 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chrX_-_154371210 | 0.45 |
ENST00000369856.8
ENST00000422373.6 ENST00000360319.9 |
FLNA
|
filamin A |
| chr6_+_37929959 | 0.44 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr12_+_8157034 | 0.44 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr12_+_20810698 | 0.44 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr3_-_197226351 | 0.43 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr7_-_111392915 | 0.43 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr10_-_28334439 | 0.42 |
ENST00000441595.2
|
MPP7
|
membrane palmitoylated protein 7 |
| chr1_+_192158448 | 0.42 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr7_+_144000320 | 0.42 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr4_-_89029881 | 0.41 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A |
| chr10_+_4786624 | 0.41 |
ENST00000533295.5
|
AKR1E2
|
aldo-keto reductase family 1 member E2 |
| chr7_+_90469634 | 0.40 |
ENST00000509356.2
|
PTTG1IP2
|
PTTG1IP family member 2 |
| chr10_+_94683722 | 0.39 |
ENST00000285979.11
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr9_-_101383558 | 0.39 |
ENST00000674556.1
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase |
| chr12_-_7503841 | 0.39 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr8_+_61287950 | 0.37 |
ENST00000519846.5
ENST00000325897.5 ENST00000523868.2 ENST00000518592.5 |
CLVS1
|
clavesin 1 |
| chr6_-_130970428 | 0.37 |
ENST00000529208.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr12_-_91178520 | 0.35 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr4_-_76023489 | 0.35 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr12_-_91153149 | 0.35 |
ENST00000550758.1
|
DCN
|
decorin |
| chrX_+_15790446 | 0.35 |
ENST00000380308.7
ENST00000307771.8 |
ZRSR2
|
zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2 |
| chr12_-_47771029 | 0.35 |
ENST00000549151.5
ENST00000548919.5 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr2_+_24049705 | 0.35 |
ENST00000380986.9
ENST00000452109.1 |
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr7_+_133253064 | 0.34 |
ENST00000393161.6
ENST00000253861.5 |
EXOC4
|
exocyst complex component 4 |
| chr2_+_209580024 | 0.34 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr2_+_24049673 | 0.33 |
ENST00000380991.8
|
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr12_-_7503744 | 0.33 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr6_+_20534441 | 0.33 |
ENST00000274695.8
ENST00000613575.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1 like 1 |
| chr2_-_213151590 | 0.33 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr6_-_130222813 | 0.32 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr4_+_40196907 | 0.32 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr9_-_92424427 | 0.31 |
ENST00000375550.5
|
OMD
|
osteomodulin |
| chr3_-_116444983 | 0.31 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr3_-_128487916 | 0.31 |
ENST00000430265.6
|
GATA2
|
GATA binding protein 2 |
| chr5_+_31193678 | 0.31 |
ENST00000265071.3
|
CDH6
|
cadherin 6 |
| chr3_-_127736329 | 0.30 |
ENST00000398101.7
|
MGLL
|
monoglyceride lipase |
| chr13_+_31739520 | 0.30 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr9_-_4666495 | 0.30 |
ENST00000475086.5
|
SPATA6L
|
spermatogenesis associated 6 like |
| chr1_-_75932392 | 0.30 |
ENST00000284142.7
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr6_+_121435595 | 0.30 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr12_-_91180365 | 0.29 |
ENST00000547937.5
|
DCN
|
decorin |
| chr7_-_23347704 | 0.29 |
ENST00000619562.4
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr15_+_41256907 | 0.29 |
ENST00000560965.1
|
CHP1
|
calcineurin like EF-hand protein 1 |
| chr17_-_40782544 | 0.28 |
ENST00000301656.4
|
KRT27
|
keratin 27 |
| chr1_+_74235377 | 0.28 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr2_-_208146150 | 0.28 |
ENST00000260988.5
|
CRYGB
|
crystallin gamma B |
| chr12_+_18738102 | 0.28 |
ENST00000317658.5
|
CAPZA3
|
capping actin protein of muscle Z-line subunit alpha 3 |
| chr10_+_94683771 | 0.27 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr7_+_95485898 | 0.27 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr13_+_31739542 | 0.27 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr11_+_6863057 | 0.27 |
ENST00000641461.1
|
OR10A2
|
olfactory receptor family 10 subfamily A member 2 |
| chr12_-_76423256 | 0.27 |
ENST00000546946.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr3_+_186717348 | 0.27 |
ENST00000447445.1
ENST00000287611.8 ENST00000644859.2 |
KNG1
|
kininogen 1 |
| chr15_+_40807055 | 0.27 |
ENST00000570108.5
ENST00000564258.5 ENST00000355341.8 ENST00000336455.9 |
ZFYVE19
|
zinc finger FYVE-type containing 19 |
| chr12_-_91182784 | 0.27 |
ENST00000547568.6
ENST00000052754.10 ENST00000552962.5 |
DCN
|
decorin |
| chr8_-_100706931 | 0.26 |
ENST00000520868.5
|
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr6_+_42050876 | 0.26 |
ENST00000465926.5
ENST00000482432.1 |
TAF8
|
TATA-box binding protein associated factor 8 |
| chr11_-_26572102 | 0.26 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr9_+_34329545 | 0.26 |
ENST00000379158.7
|
NUDT2
|
nudix hydrolase 2 |
| chr4_+_99574812 | 0.26 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr2_+_87338511 | 0.25 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr14_+_66824439 | 0.25 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr11_-_13496018 | 0.25 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr8_-_130386864 | 0.25 |
ENST00000521426.5
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr1_+_61952283 | 0.25 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chrX_+_12906612 | 0.25 |
ENST00000218032.7
|
TLR8
|
toll like receptor 8 |
| chr4_+_40197023 | 0.25 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr2_+_90234809 | 0.25 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr13_-_36297838 | 0.25 |
ENST00000511166.1
|
CCDC169-SOHLH2
|
CCDC169-SOHLH2 readthrough |
| chr17_+_47522931 | 0.24 |
ENST00000525007.5
ENST00000530173.6 |
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr14_-_74875998 | 0.24 |
ENST00000556489.4
ENST00000673765.1 |
PROX2
|
prospero homeobox 2 |
| chr9_+_34329495 | 0.24 |
ENST00000346365.8
ENST00000379155.9 ENST00000618590.1 |
NUDT2
|
nudix hydrolase 2 |
| chr2_+_90172802 | 0.24 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr11_+_118077067 | 0.24 |
ENST00000522307.5
ENST00000523251.5 ENST00000437212.8 ENST00000522824.5 ENST00000522151.5 |
TMPRSS4
|
transmembrane serine protease 4 |
| chr15_+_71096941 | 0.24 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr17_+_75721327 | 0.24 |
ENST00000579662.5
|
ITGB4
|
integrin subunit beta 4 |
| chr5_+_181040260 | 0.23 |
ENST00000515271.1
ENST00000327705.14 |
BTNL9
|
butyrophilin like 9 |
| chr2_-_189179754 | 0.23 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr11_+_118077009 | 0.23 |
ENST00000616579.4
ENST00000534111.5 |
TMPRSS4
|
transmembrane serine protease 4 |
| chr13_+_75804169 | 0.23 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr7_-_6009072 | 0.23 |
ENST00000642456.1
ENST00000642292.1 ENST00000441476.6 |
PMS2
|
PMS1 homolog 2, mismatch repair system component |
| chr2_-_88979016 | 0.22 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr17_-_40755328 | 0.22 |
ENST00000312150.5
|
KRT25
|
keratin 25 |
| chr12_+_53097656 | 0.22 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr2_+_1414382 | 0.22 |
ENST00000423320.5
ENST00000346956.7 ENST00000382198.5 |
TPO
|
thyroid peroxidase |
| chr3_-_37174578 | 0.22 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr1_-_235650748 | 0.22 |
ENST00000450593.5
ENST00000366598.8 |
GNG4
|
G protein subunit gamma 4 |
| chr19_-_57974527 | 0.21 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr3_-_139539577 | 0.21 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1 |
| chr11_+_57763820 | 0.21 |
ENST00000674106.1
|
CTNND1
|
catenin delta 1 |
| chr19_-_4558417 | 0.21 |
ENST00000586965.1
|
SEMA6B
|
semaphorin 6B |
| chr4_-_83114715 | 0.21 |
ENST00000426923.2
ENST00000311507.9 ENST00000509973.5 |
PLAC8
|
placenta associated 8 |
| chr7_+_138076453 | 0.21 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chrX_+_12906639 | 0.20 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8 |
| chr9_-_4666347 | 0.20 |
ENST00000381890.9
ENST00000682582.1 |
SPATA6L
|
spermatogenesis associated 6 like |
| chr2_+_167248638 | 0.20 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr2_+_233681877 | 0.20 |
ENST00000373426.4
|
UGT1A7
|
UDP glucuronosyltransferase family 1 member A7 |
| chr7_+_106865474 | 0.20 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr14_+_51489112 | 0.20 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chrX_-_149505274 | 0.20 |
ENST00000428056.6
ENST00000340855.11 ENST00000370441.8 |
IDS
|
iduronate 2-sulfatase |
| chr12_+_55362975 | 0.19 |
ENST00000641576.1
|
OR6C75
|
olfactory receptor family 6 subfamily C member 75 |
| chr12_+_21372899 | 0.19 |
ENST00000240652.8
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr11_-_13495984 | 0.19 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr7_-_122702912 | 0.19 |
ENST00000447240.1
ENST00000434824.2 |
RNF148
|
ring finger protein 148 |
| chr3_+_130560334 | 0.19 |
ENST00000358511.10
|
COL6A6
|
collagen type VI alpha 6 chain |
| chr1_+_84181630 | 0.19 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_-_202870761 | 0.18 |
ENST00000420558.5
ENST00000418208.5 |
ICA1L
|
islet cell autoantigen 1 like |
| chrX_-_41665766 | 0.18 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr4_-_122456725 | 0.18 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr9_-_70414657 | 0.18 |
ENST00000377126.4
|
KLF9
|
Kruppel like factor 9 |
| chr2_+_137964279 | 0.18 |
ENST00000329366.8
|
HNMT
|
histamine N-methyltransferase |
| chrX_+_37780049 | 0.18 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr3_+_109136707 | 0.18 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr1_-_241999091 | 0.18 |
ENST00000357246.4
|
MAP1LC3C
|
microtubule associated protein 1 light chain 3 gamma |
| chr11_-_18236795 | 0.18 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr5_+_31193739 | 0.18 |
ENST00000514738.5
|
CDH6
|
cadherin 6 |
| chr3_+_141262614 | 0.18 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr5_-_147906530 | 0.18 |
ENST00000318315.5
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr6_-_26250625 | 0.18 |
ENST00000618052.2
|
H3C7
|
H3 clustered histone 7 |
| chr15_-_55270280 | 0.18 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chrX_-_155264471 | 0.17 |
ENST00000369454.4
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr2_+_114442616 | 0.17 |
ENST00000410059.6
|
DPP10
|
dipeptidyl peptidase like 10 |
| chr17_-_82698637 | 0.17 |
ENST00000538809.6
ENST00000269347.10 ENST00000571995.6 |
RAB40B
|
RAB40B, member RAS oncogene family |
| chr7_-_6009019 | 0.17 |
ENST00000382321.5
ENST00000265849.12 |
PMS2
|
PMS1 homolog 2, mismatch repair system component |
| chr11_-_104898670 | 0.17 |
ENST00000422698.6
|
CASP12
|
caspase 12 (gene/pseudogene) |
| chr6_+_116461364 | 0.17 |
ENST00000368606.7
ENST00000368605.3 |
CALHM6
|
calcium homeostasis modulator family member 6 |
| chr17_-_68955332 | 0.17 |
ENST00000269080.6
ENST00000615593.4 ENST00000586539.6 ENST00000430352.6 |
ABCA8
|
ATP binding cassette subfamily A member 8 |
| chr6_-_2750912 | 0.17 |
ENST00000274643.9
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr1_+_161721563 | 0.17 |
ENST00000367948.6
|
FCRLB
|
Fc receptor like B |
| chr10_-_103153609 | 0.17 |
ENST00000675985.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr7_+_152074292 | 0.16 |
ENST00000434507.5
|
GALNT11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr19_+_15793951 | 0.16 |
ENST00000308940.8
|
OR10H5
|
olfactory receptor family 10 subfamily H member 5 |
| chr5_-_64768619 | 0.16 |
ENST00000513458.9
|
SREK1IP1
|
SREK1 interacting protein 1 |
| chr12_-_10172117 | 0.16 |
ENST00000545927.5
ENST00000309539.8 ENST00000432556.6 ENST00000544577.5 |
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr17_-_3063607 | 0.16 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor family 1 subfamily D member 5 |
| chr17_-_4967790 | 0.16 |
ENST00000575142.5
ENST00000206020.8 |
SPAG7
|
sperm associated antigen 7 |
| chr2_-_164842140 | 0.16 |
ENST00000496396.1
ENST00000629362.2 ENST00000445474.2 ENST00000483743.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr4_+_154563003 | 0.16 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chrX_-_50200988 | 0.16 |
ENST00000358526.7
|
AKAP4
|
A-kinase anchoring protein 4 |
| chr6_+_160702238 | 0.16 |
ENST00000366924.6
ENST00000308192.14 ENST00000418964.1 |
PLG
|
plasminogen |
| chr11_-_64879709 | 0.16 |
ENST00000621096.4
|
EHD1
|
EH domain containing 1 |
| chr3_+_44625027 | 0.15 |
ENST00000344387.9
ENST00000383745.6 |
ZNF197
|
zinc finger protein 197 |
| chr7_-_93226449 | 0.15 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr14_+_23630109 | 0.15 |
ENST00000432832.6
|
DHRS2
|
dehydrogenase/reductase 2 |
| chr1_+_198638968 | 0.15 |
ENST00000348564.11
ENST00000530727.5 ENST00000442510.8 ENST00000645247.1 ENST00000367367.8 ENST00000367364.5 ENST00000413409.6 |
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr16_-_84617547 | 0.15 |
ENST00000567786.2
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr7_+_95485934 | 0.15 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr7_-_28180735 | 0.15 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr6_+_150866333 | 0.15 |
ENST00000618312.4
ENST00000423867.2 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr6_+_7726089 | 0.14 |
ENST00000283147.7
|
BMP6
|
bone morphogenetic protein 6 |
| chr6_-_110815408 | 0.14 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19 |
| chr12_-_18738006 | 0.14 |
ENST00000266505.12
ENST00000543242.5 ENST00000539072.5 ENST00000541966.5 ENST00000648272.1 |
PLCZ1
|
phospholipase C zeta 1 |
| chr1_+_248095184 | 0.14 |
ENST00000358120.3
ENST00000641893.1 |
OR2L13
|
olfactory receptor family 2 subfamily L member 13 |
| chr18_+_44680093 | 0.14 |
ENST00000426838.8
ENST00000677068.1 |
SETBP1
|
SET binding protein 1 |
| chr20_-_17558811 | 0.14 |
ENST00000536626.7
ENST00000377868.6 |
BFSP1
|
beaded filament structural protein 1 |
| chr11_-_64879675 | 0.14 |
ENST00000359393.6
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH domain containing 1 |
| chr7_-_155510158 | 0.14 |
ENST00000682997.1
|
CNPY1
|
canopy FGF signaling regulator 1 |
| chr1_+_153416517 | 0.14 |
ENST00000368729.9
|
S100A7A
|
S100 calcium binding protein A7A |
| chr19_+_15728024 | 0.14 |
ENST00000305899.5
|
OR10H2
|
olfactory receptor family 10 subfamily H member 2 |
| chr5_+_157180816 | 0.14 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr12_-_9869345 | 0.14 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr7_-_107803215 | 0.13 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr18_+_616711 | 0.13 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr2_-_210476687 | 0.13 |
ENST00000233714.8
ENST00000443314.5 ENST00000441020.7 ENST00000450366.7 ENST00000431941.6 |
LANCL1
|
LanC like 1 |
| chr7_+_134866831 | 0.13 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr12_-_118359639 | 0.13 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr3_-_108529322 | 0.13 |
ENST00000273353.4
|
MYH15
|
myosin heavy chain 15 |
| chr12_-_9999176 | 0.13 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr6_+_138773747 | 0.13 |
ENST00000617445.5
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr10_-_93482194 | 0.13 |
ENST00000358334.9
ENST00000371488.3 |
MYOF
|
myoferlin |
| chr8_+_35235467 | 0.13 |
ENST00000404895.7
|
UNC5D
|
unc-5 netrin receptor D |
| chr5_-_78294656 | 0.13 |
ENST00000519295.5
ENST00000255194.11 |
AP3B1
|
adaptor related protein complex 3 subunit beta 1 |
| chrX_-_139642889 | 0.13 |
ENST00000370576.9
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr12_-_50222694 | 0.12 |
ENST00000552783.5
|
LIMA1
|
LIM domain and actin binding 1 |
| chr3_+_98130721 | 0.12 |
ENST00000641874.1
|
OR5H1
|
olfactory receptor family 5 subfamily H member 1 |
| chr14_+_22304051 | 0.12 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr3_-_20186127 | 0.12 |
ENST00000425061.5
ENST00000443724.5 ENST00000421451.5 ENST00000452020.5 ENST00000417364.1 ENST00000306698.6 ENST00000419233.6 ENST00000263753.8 ENST00000437051.5 ENST00000442720.5 ENST00000412997.6 |
SGO1
|
shugoshin 1 |
| chr2_-_99254281 | 0.12 |
ENST00000409238.5
ENST00000423800.5 |
LYG2
|
lysozyme g2 |
| chr10_-_75109085 | 0.12 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr8_-_100722731 | 0.12 |
ENST00000521865.6
ENST00000520804.2 ENST00000522720.2 ENST00000521067.1 |
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr13_-_40982880 | 0.12 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr7_-_87713287 | 0.12 |
ENST00000416177.1
ENST00000265724.8 ENST00000543898.5 |
ABCB1
|
ATP binding cassette subfamily B member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.5 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.5 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.3 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.5 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.7 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.3 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0032252 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.4 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0009804 | coumarin metabolic process(GO:0009804) flavone metabolic process(GO:0051552) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0022018 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.0 | 1.1 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.3 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.3 | GO:0006776 | retinoic acid biosynthetic process(GO:0002138) vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.6 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 2.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 0.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.4 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.1 | 1.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.3 | GO:0008940 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.2 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.0 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |