Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
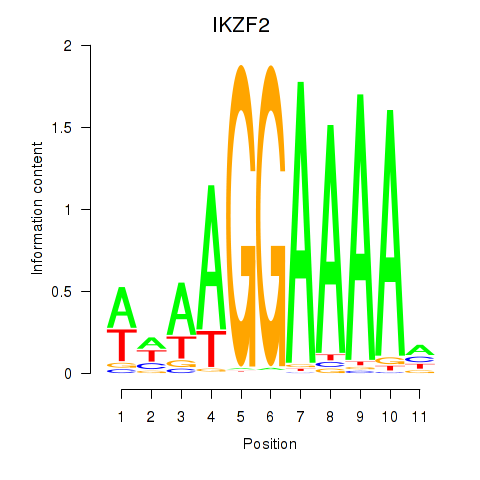
Results for IKZF2
Z-value: 0.60
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.17 | IKZF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg38_v1_chr2_-_213150236_213150334 | 0.14 | 4.7e-01 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_205422050 | 1.58 |
ENST00000367153.9
|
LEMD1
|
LEM domain containing 1 |
| chr19_+_18386150 | 1.42 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr20_+_59604527 | 1.35 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_+_209580024 | 1.25 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr12_-_121802886 | 1.21 |
ENST00000545885.5
ENST00000542933.5 ENST00000428029.6 ENST00000541694.5 ENST00000536662.5 ENST00000535643.5 ENST00000541657.5 |
LINC01089
RHOF
|
long intergenic non-protein coding RNA 1089 ras homolog family member F, filopodia associated |
| chr7_-_41700583 | 1.20 |
ENST00000442711.1
|
INHBA
|
inhibin subunit beta A |
| chrX_+_136169833 | 1.14 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_136169624 | 1.13 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1 |
| chr6_+_106086316 | 1.08 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr8_-_124565699 | 1.05 |
ENST00000519168.5
|
MTSS1
|
MTSS I-BAR domain containing 1 |
| chr2_+_209579399 | 1.01 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr15_+_45430579 | 1.01 |
ENST00000558435.5
ENST00000344300.3 ENST00000396650.7 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr2_+_209579598 | 1.01 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr7_-_23470469 | 1.01 |
ENST00000258729.8
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chrX_+_136169664 | 1.00 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chr9_+_34989641 | 0.99 |
ENST00000453597.8
ENST00000312316.9 ENST00000458263.6 ENST00000537321.5 ENST00000682809.1 ENST00000684748.1 |
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chrX_+_136148440 | 0.96 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
| chr16_+_82626955 | 0.95 |
ENST00000268613.14
ENST00000567109.6 ENST00000565636.5 ENST00000431540.7 ENST00000428848.7 |
CDH13
|
cadherin 13 |
| chr15_+_73683938 | 0.93 |
ENST00000567189.5
|
CD276
|
CD276 molecule |
| chr6_-_131063233 | 0.91 |
ENST00000392427.7
ENST00000337057.8 ENST00000525271.5 ENST00000527411.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr19_-_42765657 | 0.89 |
ENST00000406636.7
ENST00000404209.8 ENST00000306511.5 |
PSG8
|
pregnancy specific beta-1-glycoprotein 8 |
| chr6_-_131063272 | 0.88 |
ENST00000445890.6
ENST00000368128.6 ENST00000628542.2 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr2_+_157257687 | 0.88 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr2_+_102337148 | 0.85 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr2_+_9961165 | 0.83 |
ENST00000405379.6
|
GRHL1
|
grainyhead like transcription factor 1 |
| chrX_+_101550537 | 0.82 |
ENST00000372829.8
|
ARMCX1
|
armadillo repeat containing X-linked 1 |
| chrX_-_15664798 | 0.81 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator |
| chr6_+_106098933 | 0.81 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr10_-_99235783 | 0.78 |
ENST00000370546.5
ENST00000614306.1 |
HPSE2
|
heparanase 2 (inactive) |
| chr5_-_140633167 | 0.78 |
ENST00000302014.11
|
CD14
|
CD14 molecule |
| chr6_-_131063207 | 0.77 |
ENST00000530481.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr12_+_26195647 | 0.77 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr2_+_17539964 | 0.77 |
ENST00000457525.5
|
VSNL1
|
visinin like 1 |
| chr2_-_189179754 | 0.77 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chrX_+_136169891 | 0.77 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1 |
| chr17_-_17836997 | 0.74 |
ENST00000395757.6
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr7_-_122304738 | 0.73 |
ENST00000442488.7
|
FEZF1
|
FEZ family zinc finger 1 |
| chr22_-_35824373 | 0.73 |
ENST00000473487.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr5_+_96875978 | 0.70 |
ENST00000510373.5
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr3_+_12351493 | 0.70 |
ENST00000683699.1
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr9_+_72628020 | 0.70 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr3_+_130931893 | 0.69 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr2_-_24328113 | 0.68 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr17_-_41118369 | 0.64 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr5_-_151686908 | 0.64 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr5_-_147831663 | 0.64 |
ENST00000296695.10
|
SPINK1
|
serine peptidase inhibitor Kazal type 1 |
| chr4_-_48012934 | 0.63 |
ENST00000420489.7
ENST00000504722.6 |
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr17_-_676348 | 0.62 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chrX_-_41665766 | 0.60 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr7_+_93921720 | 0.56 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr12_-_27972725 | 0.55 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr5_-_147831627 | 0.55 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor Kazal type 1 |
| chr3_+_12351470 | 0.54 |
ENST00000287820.10
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr1_-_121183911 | 0.54 |
ENST00000355228.8
|
FAM72B
|
family with sequence similarity 72 member B |
| chr6_+_47698574 | 0.53 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr1_-_169630115 | 0.53 |
ENST00000263686.11
ENST00000367788.6 |
SELP
|
selectin P |
| chr19_-_43205551 | 0.52 |
ENST00000599391.1
ENST00000244295.13 ENST00000596907.5 ENST00000405312.8 ENST00000451895.1 ENST00000433626.6 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr2_+_209579429 | 0.52 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2 |
| chrX_+_50067576 | 0.51 |
ENST00000376108.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr1_-_47190013 | 0.50 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chrX_-_43882411 | 0.50 |
ENST00000378069.5
|
MAOB
|
monoamine oxidase B |
| chr15_+_69414246 | 0.50 |
ENST00000260363.9
ENST00000395392.6 |
KIF23
|
kinesin family member 23 |
| chr15_+_69414304 | 0.49 |
ENST00000352331.8
ENST00000679126.1 ENST00000647715.1 ENST00000559279.6 |
KIF23
|
kinesin family member 23 |
| chr10_-_99235846 | 0.48 |
ENST00000370552.8
ENST00000370549.5 ENST00000628193.2 |
HPSE2
|
heparanase 2 (inactive) |
| chr1_+_24319511 | 0.47 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr17_-_41055211 | 0.47 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr2_-_85410336 | 0.47 |
ENST00000263867.9
ENST00000409921.5 |
CAPG
|
capping actin protein, gelsolin like |
| chr1_+_24319342 | 0.47 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr7_-_122304499 | 0.47 |
ENST00000427185.2
|
FEZF1
|
FEZ family zinc finger 1 |
| chr18_-_49813869 | 0.46 |
ENST00000586485.5
ENST00000587994.5 ENST00000586100.1 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr19_-_42917837 | 0.45 |
ENST00000292125.6
ENST00000402603.8 ENST00000594375.1 ENST00000187910.7 |
PSG6
|
pregnancy specific beta-1-glycoprotein 6 |
| chr4_-_122456725 | 0.45 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr11_-_86068743 | 0.44 |
ENST00000356360.9
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_35122660 | 0.44 |
ENST00000603544.5
|
PRORP
|
protein only RNase P catalytic subunit |
| chr15_-_59372863 | 0.44 |
ENST00000288235.9
|
MYO1E
|
myosin IE |
| chr6_-_42217845 | 0.44 |
ENST00000053468.4
|
MRPS10
|
mitochondrial ribosomal protein S10 |
| chr17_-_17836973 | 0.43 |
ENST00000261646.11
ENST00000355815.8 |
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr6_+_27138588 | 0.43 |
ENST00000615353.1
|
H4C9
|
H4 clustered histone 9 |
| chr9_+_706841 | 0.42 |
ENST00000382293.7
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr7_+_129375643 | 0.42 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr19_-_41353904 | 0.42 |
ENST00000221930.6
|
TGFB1
|
transforming growth factor beta 1 |
| chr16_-_18876305 | 0.42 |
ENST00000563235.5
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr19_+_11089446 | 0.42 |
ENST00000557933.5
ENST00000455727.6 ENST00000535915.5 ENST00000545707.5 ENST00000558518.6 ENST00000558013.5 |
LDLR
|
low density lipoprotein receptor |
| chrX_-_64205680 | 0.41 |
ENST00000374869.8
|
AMER1
|
APC membrane recruitment protein 1 |
| chr1_+_40161355 | 0.41 |
ENST00000372771.5
|
RLF
|
RLF zinc finger |
| chr19_-_49361475 | 0.41 |
ENST00000598810.5
|
TEAD2
|
TEA domain transcription factor 2 |
| chr2_+_191678122 | 0.40 |
ENST00000425611.9
ENST00000410026.7 |
NABP1
|
nucleic acid binding protein 1 |
| chr19_-_43527189 | 0.40 |
ENST00000292147.7
ENST00000600651.5 |
ETHE1
|
ETHE1 persulfide dioxygenase |
| chr1_-_28058087 | 0.40 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr20_+_45881218 | 0.40 |
ENST00000372523.1
|
ZSWIM1
|
zinc finger SWIM-type containing 1 |
| chr2_-_133568393 | 0.40 |
ENST00000317721.10
ENST00000405974.7 ENST00000409261.6 ENST00000409213.5 |
NCKAP5
|
NCK associated protein 5 |
| chr9_-_91423819 | 0.39 |
ENST00000297689.4
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr2_-_55334529 | 0.39 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chrX_-_101407893 | 0.38 |
ENST00000676156.1
ENST00000675592.1 ENST00000674634.2 ENST00000649178.1 ENST00000218516.4 |
GLA
|
galactosidase alpha |
| chr19_-_42937098 | 0.37 |
ENST00000623675.3
ENST00000446844.3 |
PSG7
|
pregnancy specific beta-1-glycoprotein 7 |
| chr19_-_42740384 | 0.37 |
ENST00000614582.1
ENST00000595140.5 ENST00000327495.10 |
PSG3
|
pregnancy specific beta-1-glycoprotein 3 |
| chr4_-_39032922 | 0.37 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr12_-_79935069 | 0.37 |
ENST00000450142.7
|
PPP1R12A
|
protein phosphatase 1 regulatory subunit 12A |
| chr4_+_157220691 | 0.37 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr2_-_24360299 | 0.36 |
ENST00000361999.7
|
ITSN2
|
intersectin 2 |
| chr4_-_152382522 | 0.36 |
ENST00000296555.11
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr15_-_55249029 | 0.36 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr13_-_37598750 | 0.35 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr2_-_213150236 | 0.35 |
ENST00000442445.1
ENST00000342002.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr1_-_155978427 | 0.35 |
ENST00000313667.8
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr7_+_1086800 | 0.35 |
ENST00000413368.5
ENST00000397092.5 ENST00000297469.3 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr11_-_55936400 | 0.34 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr4_-_70666492 | 0.34 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr5_-_102499008 | 0.34 |
ENST00000389019.7
|
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr5_-_102498913 | 0.34 |
ENST00000513675.1
ENST00000379807.7 ENST00000506729.6 |
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr1_+_156893678 | 0.34 |
ENST00000292357.8
ENST00000338302.7 ENST00000455314.5 |
PEAR1
|
platelet endothelial aggregation receptor 1 |
| chr12_+_26195543 | 0.34 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr12_-_11134644 | 0.33 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr22_+_39901075 | 0.33 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2 |
| chr10_+_30434021 | 0.33 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr6_+_15246054 | 0.33 |
ENST00000341776.7
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr2_-_118847638 | 0.33 |
ENST00000295206.7
|
EN1
|
engrailed homeobox 1 |
| chr8_+_30387064 | 0.32 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr12_+_26195313 | 0.32 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr8_+_39913881 | 0.32 |
ENST00000518237.6
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr6_-_10412367 | 0.32 |
ENST00000379608.9
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr1_-_155978524 | 0.32 |
ENST00000361247.9
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr11_-_1757452 | 0.32 |
ENST00000427721.3
|
ENSG00000250644.3
|
novel protein |
| chr20_-_52191697 | 0.32 |
ENST00000361387.6
|
ZFP64
|
ZFP64 zinc finger protein |
| chr1_+_100719734 | 0.32 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr4_+_157220654 | 0.31 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr2_+_233032672 | 0.31 |
ENST00000233840.3
|
NEU2
|
neuraminidase 2 |
| chr6_+_116399395 | 0.31 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr1_+_117420597 | 0.31 |
ENST00000449370.6
|
MAN1A2
|
mannosidase alpha class 1A member 2 |
| chr10_+_30434176 | 0.31 |
ENST00000263056.6
ENST00000375322.2 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr3_+_179148341 | 0.30 |
ENST00000263967.4
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr14_-_56816693 | 0.30 |
ENST00000673035.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr19_+_30372364 | 0.29 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536 |
| chr11_+_18396266 | 0.29 |
ENST00000540430.5
ENST00000379412.9 |
LDHA
|
lactate dehydrogenase A |
| chr10_-_75109085 | 0.29 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr10_-_11532275 | 0.29 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr4_-_113979635 | 0.28 |
ENST00000315366.8
|
ARSJ
|
arylsulfatase family member J |
| chr16_-_58295019 | 0.28 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr16_-_58294976 | 0.28 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chr15_+_66386902 | 0.28 |
ENST00000307102.10
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr2_+_26970628 | 0.28 |
ENST00000233121.7
ENST00000405074.7 |
MAPRE3
|
microtubule associated protein RP/EB family member 3 |
| chr10_-_101839818 | 0.28 |
ENST00000348850.9
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr10_+_119818699 | 0.27 |
ENST00000650409.1
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr22_-_28711931 | 0.27 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr1_+_161749762 | 0.27 |
ENST00000367943.5
|
DUSP12
|
dual specificity phosphatase 12 |
| chr8_+_96584920 | 0.27 |
ENST00000521590.5
|
SDC2
|
syndecan 2 |
| chr22_-_35840218 | 0.27 |
ENST00000414461.6
ENST00000416721.6 ENST00000449924.6 ENST00000262829.11 ENST00000397305.3 |
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr13_+_27620945 | 0.27 |
ENST00000621089.2
|
POLR1D
|
RNA polymerase I and III subunit D |
| chr1_+_158831323 | 0.26 |
ENST00000368141.5
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr4_+_133149307 | 0.26 |
ENST00000618019.1
|
PCDH10
|
protocadherin 10 |
| chr15_+_81000913 | 0.26 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chr4_+_147732070 | 0.26 |
ENST00000336498.8
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr7_+_107583919 | 0.26 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr17_+_7484357 | 0.25 |
ENST00000674977.2
|
POLR2A
|
RNA polymerase II subunit A |
| chr2_+_135531460 | 0.25 |
ENST00000683871.1
ENST00000409478.5 ENST00000264160.8 ENST00000438014.5 |
R3HDM1
|
R3H domain containing 1 |
| chr7_+_120988683 | 0.25 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr3_+_10026409 | 0.25 |
ENST00000287647.7
ENST00000676013.1 ENST00000675286.1 ENST00000419585.5 |
FANCD2
|
FA complementation group D2 |
| chr12_+_59664677 | 0.25 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr12_-_10986912 | 0.25 |
ENST00000506868.1
|
TAS2R50
|
taste 2 receptor member 50 |
| chr5_+_58491451 | 0.25 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr22_-_35840577 | 0.25 |
ENST00000405409.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr12_+_6494087 | 0.25 |
ENST00000382457.8
ENST00000315579.10 |
NCAPD2
|
non-SMC condensin I complex subunit D2 |
| chr5_-_152405277 | 0.24 |
ENST00000255262.4
|
NMUR2
|
neuromedin U receptor 2 |
| chr2_+_188974364 | 0.24 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr16_-_20900319 | 0.24 |
ENST00000564349.5
ENST00000324344.9 |
ERI2
DCUN1D3
|
ERI1 exoribonuclease family member 2 defective in cullin neddylation 1 domain containing 3 |
| chr18_+_24460630 | 0.24 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4 |
| chr11_+_22338333 | 0.24 |
ENST00000263160.4
|
SLC17A6
|
solute carrier family 17 member 6 |
| chr1_-_152325232 | 0.24 |
ENST00000368799.2
|
FLG
|
filaggrin |
| chr14_+_35122722 | 0.24 |
ENST00000605870.5
ENST00000557404.3 |
PRORP
|
protein only RNase P catalytic subunit |
| chr10_-_75109172 | 0.24 |
ENST00000372700.7
ENST00000473072.2 ENST00000491677.6 ENST00000372702.7 |
DUSP13
|
dual specificity phosphatase 13 |
| chr11_+_126283059 | 0.24 |
ENST00000392679.6
ENST00000392678.7 ENST00000392680.6 |
TIRAP
|
TIR domain containing adaptor protein |
| chr11_-_72674394 | 0.23 |
ENST00000418754.6
ENST00000334456.10 ENST00000542969.2 |
PDE2A
|
phosphodiesterase 2A |
| chr10_-_96271508 | 0.23 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr7_-_106284524 | 0.23 |
ENST00000681936.1
ENST00000680786.1 ENST00000681550.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr7_-_14841267 | 0.23 |
ENST00000406247.7
ENST00000399322.7 |
DGKB
|
diacylglycerol kinase beta |
| chr12_-_55688891 | 0.23 |
ENST00000557555.2
|
ITGA7
|
integrin subunit alpha 7 |
| chr13_-_46182136 | 0.23 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr11_-_19241598 | 0.23 |
ENST00000532666.1
ENST00000527884.5 ENST00000620009.4 |
E2F8
|
E2F transcription factor 8 |
| chr10_-_96271553 | 0.22 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr21_+_38256984 | 0.22 |
ENST00000398938.7
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr18_+_24460655 | 0.22 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chrX_+_28587411 | 0.22 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chr14_-_106211453 | 0.21 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr17_+_7012417 | 0.21 |
ENST00000548577.5
|
RNASEK
|
ribonuclease K |
| chr12_+_106582996 | 0.21 |
ENST00000392842.6
|
RFX4
|
regulatory factor X4 |
| chr20_-_14337602 | 0.21 |
ENST00000378053.3
ENST00000341420.5 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr12_-_104050112 | 0.21 |
ENST00000547583.1
ENST00000546851.1 ENST00000360814.9 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr3_-_169769542 | 0.21 |
ENST00000330368.3
|
ACTRT3
|
actin related protein T3 |
| chr12_-_91179517 | 0.21 |
ENST00000551354.1
|
DCN
|
decorin |
| chr9_+_136807911 | 0.20 |
ENST00000371671.9
ENST00000311502.12 ENST00000371663.10 |
RABL6
|
RAB, member RAS oncogene family like 6 |
| chr19_+_18340581 | 0.20 |
ENST00000604499.6
ENST00000269919.11 ENST00000595066.5 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr6_+_142147162 | 0.20 |
ENST00000452973.6
ENST00000620996.4 ENST00000367621.1 ENST00000367630.9 |
VTA1
|
vesicle trafficking 1 |
| chr2_+_196639686 | 0.20 |
ENST00000389175.9
|
CCDC150
|
coiled-coil domain containing 150 |
| chr17_-_40782544 | 0.20 |
ENST00000301656.4
|
KRT27
|
keratin 27 |
| chr19_+_49930219 | 0.20 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr2_+_48568981 | 0.19 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr17_-_38748184 | 0.19 |
ENST00000618941.4
ENST00000620225.5 ENST00000618506.1 ENST00000616129.4 |
PCGF2
|
polycomb group ring finger 2 |
| chr1_-_150269051 | 0.19 |
ENST00000414276.6
ENST00000360244.8 |
APH1A
|
aph-1 homolog A, gamma-secretase subunit |
| chr4_+_176065980 | 0.19 |
ENST00000280190.8
|
WDR17
|
WD repeat domain 17 |
| chr6_-_11778781 | 0.19 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.4 | 1.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.2 | GO:0071221 | response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.2 | 1.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 1.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.2 | 1.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.8 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 1.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 2.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.4 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.4 | GO:0052251 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.4 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.4 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.9 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.3 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.1 | 0.6 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.3 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 1.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.3 | GO:0003409 | optic vesicle morphogenesis(GO:0003404) optic cup structural organization(GO:0003409) |
| 0.1 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.4 | GO:1900223 | positive regulation of beta-amyloid clearance(GO:1900223) |
| 0.1 | 0.5 | GO:0014063 | response to aluminum ion(GO:0010044) negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 1.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.3 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.6 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.9 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.5 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 4.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 1.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:1904170 | positive regulation of interleukin-1 alpha secretion(GO:0050717) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.3 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 3.8 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.3 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.4 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.0 | GO:0060100 | positive regulation of phagocytosis, engulfment(GO:0060100) positive regulation of membrane invagination(GO:1905155) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.2 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.3 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.0 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 1.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 1.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 2.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.3 | GO:0071754 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 1.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.3 | 0.9 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.3 | 0.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 1.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 2.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 4.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0051990 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.0 | 0.3 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 3.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 4.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 3.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |