Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for IRF7
Z-value: 0.96
Transcription factors associated with IRF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF7
|
ENSG00000185507.21 | IRF7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF7 | hg38_v1_chr11_-_615570_615728 | -0.10 | 5.9e-01 | Click! |
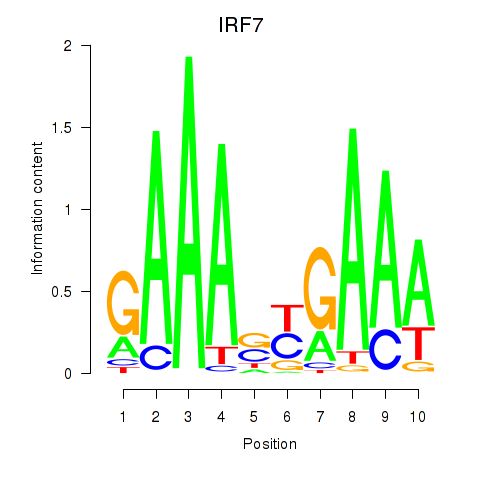
Activity profile of IRF7 motif
Sorted Z-values of IRF7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.9 | 2.8 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.6 | 1.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.6 | 1.9 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.4 | 3.1 | GO:0030421 | defecation(GO:0030421) |
| 0.4 | 1.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 2.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.3 | 0.3 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.3 | 1.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.3 | 0.9 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.3 | 1.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.3 | 1.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.3 | 2.3 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.3 | 2.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.7 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 1.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 2.5 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 2.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.2 | 3.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 1.0 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 0.6 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.8 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 4.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 1.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 0.6 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.2 | 0.6 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 0.6 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.2 | 0.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 0.5 | GO:1900238 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.2 | 2.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.7 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.4 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.1 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.7 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 1.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.4 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.4 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.7 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.3 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 0.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.5 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 1.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 1.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.4 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 2.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 1.4 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.3 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 0.8 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 0.5 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 1.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.4 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.3 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 1.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.4 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 1.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.7 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 1.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 1.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.2 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) regulation of amacrine cell differentiation(GO:1902869) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.1 | GO:1903781 | positive regulation of cardiac conduction(GO:1903781) |
| 0.1 | 0.3 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.1 | 0.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0021886 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) sensory system development(GO:0048880) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.2 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.1 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.5 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.6 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.3 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.2 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:1905154 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.7 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.4 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 1.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.6 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.7 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.5 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.2 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.8 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.8 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 2.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 1.8 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.5 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0060932 | sinoatrial node cell development(GO:0060931) His-Purkinje system cell differentiation(GO:0060932) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 1.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.7 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.3 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.8 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) |
| 0.0 | 0.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.3 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.5 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.3 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 1.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 3.6 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 6.8 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.0 | GO:0042088 | T-helper 1 type immune response(GO:0042088) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.1 | GO:1900376 | regulation of melanin biosynthetic process(GO:0048021) positive regulation of melanin biosynthetic process(GO:0048023) regulation of secondary metabolite biosynthetic process(GO:1900376) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0046476 | glucosylceramide biosynthetic process(GO:0006679) glycosylceramide biosynthetic process(GO:0046476) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.4 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:1901895 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 1.1 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.4 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.7 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.3 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.2 | GO:0051000 | positive regulation of nitric-oxide synthase activity(GO:0051000) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 0.3 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:1904261 | positive regulation of extracellular matrix assembly(GO:1901203) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 2.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 1.5 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.0 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 3.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.5 | 1.6 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.3 | 1.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.2 | 1.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 2.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 1.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 9.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.3 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 1.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 0.2 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.7 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.5 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.7 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 1.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 3.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 2.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.6 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.4 | 5.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 0.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 0.9 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.3 | 2.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 1.6 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.3 | 4.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 0.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.3 | 1.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 1.3 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.3 | 1.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.9 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.2 | 1.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 1.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 0.6 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 2.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.6 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 2.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 3.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 3.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.6 | GO:0043208 | fucose binding(GO:0042806) glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.4 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 1.0 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.5 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.6 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.1 | 0.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 1.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.5 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 1.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) cholestenol delta-isomerase activity(GO:0047750) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 1.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 5.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 5.9 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.0 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 1.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 5.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 6.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 13.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.3 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.9 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 3.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |