Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
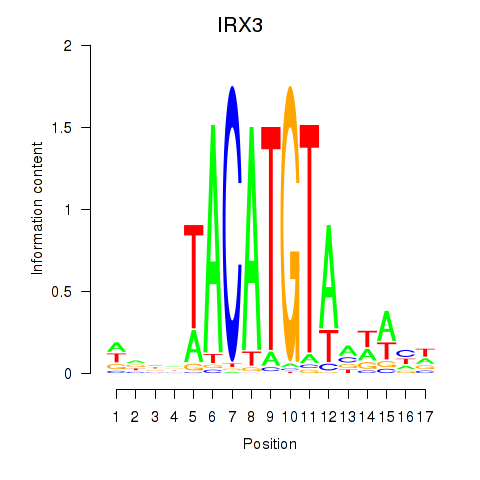
Results for IRX3
Z-value: 1.01
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.12 | IRX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg38_v1_chr16_-_54286763_54286803 | -0.11 | 5.7e-01 | Click! |
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_55833186 | 5.59 |
ENST00000361503.8
ENST00000422046.6 |
CES1
|
carboxylesterase 1 |
| chr16_-_55833085 | 3.83 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1 |
| chr4_+_68815991 | 2.67 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr4_+_69096467 | 2.39 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr4_+_69096494 | 2.31 |
ENST00000508661.5
ENST00000622664.1 |
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr10_+_116427839 | 2.15 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr3_+_319683 | 2.03 |
ENST00000620033.4
|
CHL1
|
cell adhesion molecule L1 like |
| chr2_+_89936859 | 2.01 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr10_+_116545907 | 1.91 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr12_-_91179355 | 1.80 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr12_-_70788914 | 1.78 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr4_-_69860138 | 1.65 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1 |
| chr5_+_36608146 | 1.61 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr12_-_91180365 | 1.55 |
ENST00000547937.5
|
DCN
|
decorin |
| chr12_-_10130241 | 1.52 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr19_+_41088450 | 1.42 |
ENST00000330436.4
|
CYP2A13
|
cytochrome P450 family 2 subfamily A member 13 |
| chr4_+_69995958 | 1.39 |
ENST00000381060.2
ENST00000246895.9 |
STATH
|
statherin |
| chr12_-_10130143 | 1.37 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr5_+_140841183 | 1.36 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr16_+_19410480 | 1.35 |
ENST00000541464.5
|
TMC5
|
transmembrane channel like 5 |
| chr9_+_87497222 | 1.32 |
ENST00000358077.9
|
DAPK1
|
death associated protein kinase 1 |
| chr12_-_10130082 | 1.32 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr11_+_27055215 | 1.20 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr6_+_27138588 | 1.13 |
ENST00000615353.1
|
H4C9
|
H4 clustered histone 9 |
| chr4_+_69280472 | 1.08 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
| chr5_+_140855882 | 1.07 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr12_-_91179472 | 1.05 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr7_-_99976017 | 1.04 |
ENST00000411734.1
ENST00000292401.9 |
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr22_-_18936142 | 1.01 |
ENST00000438924.5
ENST00000457083.1 ENST00000357068.11 ENST00000420436.5 ENST00000334029.6 ENST00000610940.4 |
PRODH
|
proline dehydrogenase 1 |
| chr2_-_89245596 | 0.98 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr4_-_69214743 | 0.90 |
ENST00000446444.2
|
UGT2B11
|
UDP glucuronosyltransferase family 2 member B11 |
| chr4_+_41359599 | 0.86 |
ENST00000513024.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_+_93566659 | 0.81 |
ENST00000371481.9
ENST00000371483.8 ENST00000604414.1 |
FFAR4
|
free fatty acid receptor 4 |
| chr12_-_13095664 | 0.80 |
ENST00000337630.10
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr7_-_93148345 | 0.74 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr6_-_27831557 | 0.73 |
ENST00000611927.2
|
H4C12
|
H4 clustered histone 12 |
| chr12_+_69348372 | 0.71 |
ENST00000261267.7
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr6_-_52840843 | 0.70 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr2_-_87825952 | 0.70 |
ENST00000398146.4
|
RGPD2
|
RANBP2 like and GRIP domain containing 2 |
| chr4_-_170003738 | 0.70 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr1_-_108200849 | 0.69 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 member 24 |
| chr4_-_103077282 | 0.68 |
ENST00000503230.5
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9 member B2 |
| chr3_-_74521140 | 0.68 |
ENST00000263665.6
|
CNTN3
|
contactin 3 |
| chr8_-_139654330 | 0.68 |
ENST00000647605.1
|
KCNK9
|
potassium two pore domain channel subfamily K member 9 |
| chr1_-_46132616 | 0.67 |
ENST00000423209.5
ENST00000262741.10 |
PIK3R3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr5_+_74715503 | 0.66 |
ENST00000513336.5
|
HEXB
|
hexosaminidase subunit beta |
| chrX_+_7147675 | 0.66 |
ENST00000674429.1
|
STS
|
steroid sulfatase |
| chrX_-_15602150 | 0.65 |
ENST00000427411.2
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr6_-_47042260 | 0.65 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr1_-_46132650 | 0.64 |
ENST00000372006.5
ENST00000425892.2 ENST00000420542.5 |
PIK3R3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr1_+_158289916 | 0.64 |
ENST00000368170.8
|
CD1C
|
CD1c molecule |
| chr19_+_50188180 | 0.64 |
ENST00000598205.5
|
MYH14
|
myosin heavy chain 14 |
| chr9_+_102995308 | 0.64 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr22_+_22704265 | 0.63 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr22_+_22818994 | 0.63 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 |
| chr4_+_41538143 | 0.61 |
ENST00000503057.6
ENST00000511496.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr14_-_106005574 | 0.61 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr10_+_94683771 | 0.60 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr21_-_32603237 | 0.60 |
ENST00000431599.1
|
CFAP298
|
cilia and flagella associated protein 298 |
| chr4_-_88284747 | 0.59 |
ENST00000514204.1
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr16_-_4788254 | 0.58 |
ENST00000591624.1
ENST00000268231.13 ENST00000396693.9 |
SEPTIN12
|
septin 12 |
| chr14_-_105987068 | 0.58 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr14_-_106470788 | 0.56 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr4_-_103076688 | 0.56 |
ENST00000394785.9
|
SLC9B2
|
solute carrier family 9 member B2 |
| chr4_-_87529359 | 0.56 |
ENST00000458304.2
ENST00000282470.11 |
SPARCL1
|
SPARC like 1 |
| chr6_+_29396555 | 0.55 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr10_+_29289061 | 0.55 |
ENST00000375500.8
ENST00000649382.2 |
LYZL1
|
lysozyme like 1 |
| chrX_+_7147237 | 0.54 |
ENST00000666110.2
|
STS
|
steroid sulfatase |
| chr6_+_131637296 | 0.54 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr2_+_106063234 | 0.54 |
ENST00000409944.5
|
ECRG4
|
ECRG4 augurin precursor |
| chr1_+_103617427 | 0.54 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr5_+_148063971 | 0.54 |
ENST00000398454.5
ENST00000359874.7 ENST00000508733.5 ENST00000256084.8 |
SPINK5
|
serine peptidase inhibitor Kazal type 5 |
| chr17_-_31314066 | 0.53 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr16_-_20697680 | 0.52 |
ENST00000520010.6
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr7_+_143620967 | 0.52 |
ENST00000684770.1
|
TCAF2
|
TRPM8 channel associated factor 2 |
| chr5_-_177780633 | 0.52 |
ENST00000513554.5
ENST00000440605.7 |
FAM153A
|
family with sequence similarity 153 member A |
| chr19_-_3500664 | 0.51 |
ENST00000427575.6
|
DOHH
|
deoxyhypusine hydroxylase |
| chr11_+_57597563 | 0.50 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr8_+_22367526 | 0.49 |
ENST00000289952.9
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 member 14 |
| chr14_-_106185387 | 0.49 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr2_-_207167220 | 0.49 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr22_+_50738198 | 0.48 |
ENST00000216139.10
ENST00000529621.1 |
ACR
|
acrosin |
| chr4_+_87832917 | 0.47 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr11_-_111911759 | 0.47 |
ENST00000650687.2
|
CRYAB
|
crystallin alpha B |
| chr14_+_22168387 | 0.46 |
ENST00000557168.1
|
TRAV30
|
T cell receptor alpha variable 30 |
| chr22_-_36160773 | 0.46 |
ENST00000531095.1
ENST00000349314.6 ENST00000397293.6 |
APOL3
|
apolipoprotein L3 |
| chr6_-_32953017 | 0.46 |
ENST00000395305.7
ENST00000374843.9 ENST00000395303.7 ENST00000429234.1 |
HLA-DMA
ENSG00000248993.1
|
major histocompatibility complex, class II, DM alpha novel protein |
| chr19_-_10380454 | 0.46 |
ENST00000530829.1
ENST00000529370.5 |
TYK2
|
tyrosine kinase 2 |
| chr19_+_37594830 | 0.46 |
ENST00000589117.5
|
ZNF540
|
zinc finger protein 540 |
| chr1_+_76867469 | 0.45 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr8_-_143609547 | 0.45 |
ENST00000433751.5
ENST00000495276.6 ENST00000220966.10 |
PYCR3
|
pyrroline-5-carboxylate reductase 3 |
| chr10_+_89332484 | 0.45 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr13_-_46142834 | 0.44 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr11_-_65382632 | 0.44 |
ENST00000294187.10
ENST00000398802.6 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25 member 45 |
| chr10_+_89301932 | 0.44 |
ENST00000371826.4
ENST00000679755.1 |
IFIT2
|
interferon induced protein with tetratricopeptide repeats 2 |
| chr8_-_142917843 | 0.43 |
ENST00000323110.2
|
CYP11B2
|
cytochrome P450 family 11 subfamily B member 2 |
| chr14_-_24339951 | 0.43 |
ENST00000216274.10
|
RIPK3
|
receptor interacting serine/threonine kinase 3 |
| chr2_-_182242031 | 0.43 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr12_-_10098977 | 0.43 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr14_-_94476240 | 0.43 |
ENST00000424550.6
ENST00000337425.10 ENST00000674164.1 ENST00000546329.2 |
SERPINA9
|
serpin family A member 9 |
| chr14_-_51096029 | 0.42 |
ENST00000298355.7
|
TRIM9
|
tripartite motif containing 9 |
| chr19_-_43883964 | 0.42 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr5_-_135954962 | 0.42 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr1_+_160739265 | 0.41 |
ENST00000368042.7
|
SLAMF7
|
SLAM family member 7 |
| chr2_-_232550537 | 0.41 |
ENST00000408957.7
|
TIGD1
|
tigger transposable element derived 1 |
| chr11_-_4288083 | 0.41 |
ENST00000638166.1
|
SSU72P4
|
SSU72 pseudogene 4 |
| chr20_-_13990609 | 0.41 |
ENST00000284951.10
ENST00000378072.5 |
SEL1L2
|
SEL1L2 adaptor subunit of ERAD E3 ligase |
| chr16_-_72172135 | 0.40 |
ENST00000537465.5
ENST00000237353.15 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr19_-_54364908 | 0.40 |
ENST00000391742.7
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr4_+_73740541 | 0.40 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr11_+_60280577 | 0.39 |
ENST00000679988.1
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr1_+_160739239 | 0.39 |
ENST00000368043.8
|
SLAMF7
|
SLAM family member 7 |
| chr12_+_55720405 | 0.39 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 |
| chr2_+_100974849 | 0.39 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr4_-_99657820 | 0.39 |
ENST00000511828.2
|
C4orf54
|
chromosome 4 open reading frame 54 |
| chr20_+_56358938 | 0.39 |
ENST00000371384.4
ENST00000437418.1 |
FAM210B
|
family with sequence similarity 210 member B |
| chr21_-_32813695 | 0.39 |
ENST00000479548.2
ENST00000490358.5 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr1_+_202348687 | 0.38 |
ENST00000608999.6
ENST00000391959.5 ENST00000480184.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr2_-_24084292 | 0.38 |
ENST00000413037.1
ENST00000407482.5 |
TP53I3
|
tumor protein p53 inducible protein 3 |
| chr12_+_55720367 | 0.38 |
ENST00000547072.5
ENST00000552930.5 ENST00000257895.10 |
RDH5
|
retinol dehydrogenase 5 |
| chr3_-_151329539 | 0.37 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr12_-_91153149 | 0.37 |
ENST00000550758.1
|
DCN
|
decorin |
| chr16_-_57802401 | 0.37 |
ENST00000569112.5
ENST00000445690.7 ENST00000562311.5 ENST00000379655.8 |
KIFC3
|
kinesin family member C3 |
| chr11_-_26567087 | 0.37 |
ENST00000436318.6
ENST00000281268.12 |
MUC15
|
mucin 15, cell surface associated |
| chr3_-_65622073 | 0.37 |
ENST00000621418.4
ENST00000611645.4 |
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr12_+_20810698 | 0.37 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr20_+_2295994 | 0.36 |
ENST00000381458.6
|
TGM3
|
transglutaminase 3 |
| chr6_+_121435595 | 0.36 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr15_-_21742799 | 0.36 |
ENST00000622410.2
|
ENSG00000278263.2
|
novel protein, identical to IGHV4-4 |
| chr12_+_5432101 | 0.36 |
ENST00000423158.4
|
NTF3
|
neurotrophin 3 |
| chr1_+_202462730 | 0.36 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chrX_-_42778155 | 0.35 |
ENST00000378131.4
|
PPP1R2C
|
PPP1R2C family member C |
| chr10_-_95069489 | 0.35 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chr1_-_161367872 | 0.35 |
ENST00000367974.2
|
CFAP126
|
cilia and flagella associated protein 126 |
| chr7_+_143620925 | 0.35 |
ENST00000444908.7
|
TCAF2
|
TRPM8 channel associated factor 2 |
| chr14_-_106349792 | 0.35 |
ENST00000438142.3
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr7_-_120858066 | 0.35 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr9_-_90642855 | 0.35 |
ENST00000637905.1
|
DIRAS2
|
DIRAS family GTPase 2 |
| chr12_-_90955172 | 0.35 |
ENST00000358859.3
|
CCER1
|
coiled-coil glutamate rich protein 1 |
| chr1_+_220690354 | 0.35 |
ENST00000294889.6
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr19_+_54497879 | 0.35 |
ENST00000412608.5
ENST00000610651.1 |
LAIR2
|
leukocyte associated immunoglobulin like receptor 2 |
| chr2_+_233636502 | 0.34 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase family 1 member A10 |
| chr9_-_37465402 | 0.34 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr7_-_64982021 | 0.34 |
ENST00000610793.1
ENST00000620222.4 |
ZNF117
|
zinc finger protein 117 |
| chr12_-_10453330 | 0.34 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr13_-_37598750 | 0.34 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr10_+_94938649 | 0.34 |
ENST00000461906.1
ENST00000260682.8 |
CYP2C9
|
cytochrome P450 family 2 subfamily C member 9 |
| chr16_-_67190099 | 0.34 |
ENST00000314586.11
ENST00000563889.1 ENST00000564418.1 ENST00000545725.6 |
EXOC3L1
|
exocyst complex component 3 like 1 |
| chr14_-_106012390 | 0.34 |
ENST00000455737.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr15_-_22185402 | 0.33 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr5_-_132543513 | 0.33 |
ENST00000231454.6
|
IL5
|
interleukin 5 |
| chr3_-_191282383 | 0.33 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr19_+_3762705 | 0.33 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr2_-_157444044 | 0.32 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr11_-_60183011 | 0.32 |
ENST00000533023.5
ENST00000420732.6 ENST00000528851.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr8_+_119873710 | 0.32 |
ENST00000523492.5
ENST00000286234.6 |
DEPTOR
|
DEP domain containing MTOR interacting protein |
| chr12_-_101830799 | 0.32 |
ENST00000549940.5
ENST00000392919.4 |
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta |
| chr6_+_30940970 | 0.32 |
ENST00000462446.6
ENST00000304311.3 |
MUCL3
|
mucin like 3 |
| chr19_+_3762665 | 0.32 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr12_+_21526287 | 0.32 |
ENST00000256969.7
|
SPX
|
spexin hormone |
| chr1_+_160739286 | 0.32 |
ENST00000359331.8
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr7_-_111562455 | 0.31 |
ENST00000452895.5
ENST00000405709.7 ENST00000452753.1 ENST00000331762.7 |
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr2_+_113072318 | 0.31 |
ENST00000393197.3
|
IL1F10
|
interleukin 1 family member 10 |
| chr7_+_80646305 | 0.31 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr5_+_141192330 | 0.31 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr1_+_26317950 | 0.31 |
ENST00000374213.3
|
CD52
|
CD52 molecule |
| chr12_-_9208388 | 0.31 |
ENST00000261336.7
|
PZP
|
PZP alpha-2-macroglobulin like |
| chr14_-_106025628 | 0.31 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr2_-_68952880 | 0.30 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr16_-_3443446 | 0.30 |
ENST00000301744.7
|
ZNF597
|
zinc finger protein 597 |
| chr17_-_63827647 | 0.30 |
ENST00000584574.5
ENST00000585145.1 ENST00000427159.7 |
FTSJ3
|
FtsJ RNA 2'-O-methyltransferase 3 |
| chr22_+_50486835 | 0.30 |
ENST00000216075.11
ENST00000395732.7 |
MIOX
|
myo-inositol oxygenase |
| chr17_+_3475959 | 0.30 |
ENST00000263080.3
|
ASPA
|
aspartoacylase |
| chr3_-_57292676 | 0.30 |
ENST00000389601.3
ENST00000487349.6 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr2_+_190927649 | 0.29 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr12_+_54854505 | 0.29 |
ENST00000308796.11
ENST00000619042.1 |
MUCL1
|
mucin like 1 |
| chr6_-_55579160 | 0.29 |
ENST00000370850.6
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase like 1 |
| chr1_+_162497805 | 0.29 |
ENST00000538489.5
ENST00000489294.2 |
UHMK1
|
U2AF homology motif kinase 1 |
| chr3_+_178419123 | 0.28 |
ENST00000614557.1
ENST00000455307.5 ENST00000436432.1 |
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript) long intergenic non-protein coding RNA 1014 |
| chr5_-_146922515 | 0.28 |
ENST00000508545.6
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr14_-_106811131 | 0.28 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr13_+_21140514 | 0.28 |
ENST00000382533.8
ENST00000621421.4 |
SAP18
|
Sin3A associated protein 18 |
| chr3_-_194672175 | 0.28 |
ENST00000265245.10
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr1_-_115841116 | 0.28 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2 |
| chr7_+_39566366 | 0.28 |
ENST00000448268.5
ENST00000223273.7 ENST00000432096.2 |
YAE1
|
YAE1 maturation factor of ABCE1 |
| chr10_-_35090545 | 0.28 |
ENST00000374751.7
ENST00000626172.2 |
CUL2
|
cullin 2 |
| chrX_-_34132311 | 0.28 |
ENST00000346193.5
ENST00000613251.1 |
FAM47A
|
family with sequence similarity 47 member A |
| chrX_-_6535118 | 0.28 |
ENST00000381089.7
ENST00000612369.4 ENST00000398729.1 |
VCX3A
|
variable charge X-linked 3A |
| chr4_-_144019287 | 0.28 |
ENST00000638448.1
ENST00000513128.5 ENST00000506516.6 ENST00000429670.3 ENST00000502664.6 |
GYPB
|
glycophorin B (MNS blood group) |
| chr12_-_100267055 | 0.27 |
ENST00000551642.1
ENST00000550587.6 ENST00000416321.5 ENST00000549249.5 |
DEPDC4
|
DEP domain containing 4 |
| chr19_+_58305319 | 0.27 |
ENST00000413518.5
ENST00000427361.5 ENST00000610038.5 ENST00000608070.5 ENST00000609864.5 |
ERVK3-1
|
endogenous retrovirus group K3 member 1 |
| chr19_-_23274194 | 0.27 |
ENST00000640920.1
ENST00000639997.1 ENST00000639327.1 ENST00000640517.1 ENST00000639752.1 ENST00000640354.1 ENST00000638919.1 ENST00000638822.1 ENST00000640838.1 ENST00000611392.5 ENST00000594653.1 |
ENSG00000284428.1
ENSG00000283201.1
ENSG00000269107.1
|
novel transcript zinc finger protein 724 novel transcript |
| chrX_+_12906639 | 0.27 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8 |
| chr1_+_116754422 | 0.27 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr20_-_18497218 | 0.27 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase |
| chr8_-_109680812 | 0.27 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr12_-_21334858 | 0.27 |
ENST00000445053.1
ENST00000458504.5 ENST00000422327.5 ENST00000683939.1 |
SLCO1A2
|
solute carrier organic anion transporter family member 1A2 |
| chr1_-_243843164 | 0.27 |
ENST00000491219.6
ENST00000680056.1 ENST00000492957.2 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr12_+_10212867 | 0.27 |
ENST00000545047.5
ENST00000266458.10 ENST00000629504.1 ENST00000543602.5 ENST00000545887.1 |
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr22_+_36863091 | 0.27 |
ENST00000650698.1
|
NCF4
|
neutrophil cytosolic factor 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.4 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.6 | 1.7 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.3 | 1.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 1.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.3 | 1.4 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.3 | 4.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 4.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 0.9 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 1.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.9 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 0.5 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.2 | 4.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.2 | 0.5 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.2 | 0.5 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 0.5 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.4 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.1 | 1.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.4 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 1.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.7 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.4 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 1.4 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 1.9 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 0.4 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.2 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 0.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.5 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.4 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.7 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.5 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.7 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.3 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.9 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.6 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.2 | GO:0046021 | positive regulation of transcription during mitosis(GO:0045897) regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) |
| 0.1 | 1.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 2.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 1.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 6.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.5 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 1.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.3 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.2 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.3 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.6 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.4 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.6 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.0 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.6 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0071287 | cellular response to magnesium ion(GO:0071286) cellular response to manganese ion(GO:0071287) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:1905146 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 0.8 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.2 | 0.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 2.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 11.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 2.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.4 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.5 | 1.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.4 | 1.2 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.4 | 1.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.2 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.3 | 1.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 8.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.4 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 4.5 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.7 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 3.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.5 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 1.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 2.0 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.6 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.9 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.5 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 5.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 3.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 5.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |