Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
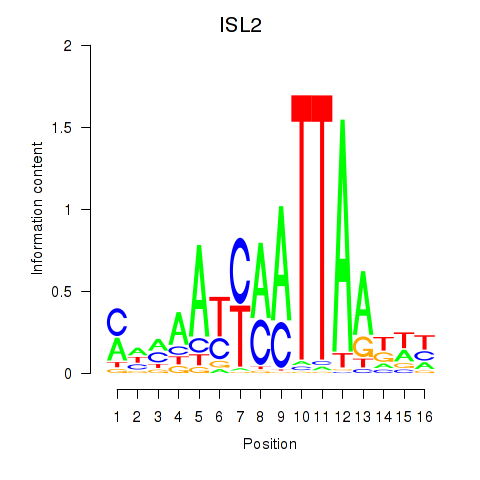
Results for ISL2
Z-value: 0.45
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.10 | ISL2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL2 | hg38_v1_chr15_+_76336755_76336779 | 0.57 | 9.2e-04 | Click! |
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_123601815 | 1.40 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_123601836 | 1.39 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr9_+_72577939 | 0.96 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr2_-_162152404 | 0.88 |
ENST00000375497.3
|
GCG
|
glucagon |
| chr1_-_247760556 | 0.77 |
ENST00000641256.1
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr5_-_140346596 | 0.75 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr12_-_95551417 | 0.73 |
ENST00000258499.8
|
USP44
|
ubiquitin specific peptidase 44 |
| chr11_-_5301946 | 0.70 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr2_-_162152239 | 0.61 |
ENST00000418842.7
|
GCG
|
glucagon |
| chr5_+_90640718 | 0.60 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr11_-_28108109 | 0.57 |
ENST00000263181.7
|
KIF18A
|
kinesin family member 18A |
| chr9_+_12693327 | 0.55 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr12_+_1970772 | 0.50 |
ENST00000682544.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr12_+_1970809 | 0.50 |
ENST00000683781.1
ENST00000682462.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr11_-_107858777 | 0.47 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr17_-_3592877 | 0.46 |
ENST00000399756.8
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chr11_-_124320197 | 0.44 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr6_+_130018565 | 0.41 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr3_-_142029108 | 0.40 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr9_+_72577788 | 0.39 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr9_+_72577369 | 0.39 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr14_+_92323154 | 0.37 |
ENST00000532405.6
ENST00000676001.1 ENST00000531433.5 |
SLC24A4
|
solute carrier family 24 member 4 |
| chr6_-_155455830 | 0.37 |
ENST00000159060.3
|
NOX3
|
NADPH oxidase 3 |
| chr15_-_82709886 | 0.37 |
ENST00000666055.1
ENST00000261722.8 ENST00000535513.2 |
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr17_-_8119047 | 0.36 |
ENST00000318227.4
|
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr4_+_94207596 | 0.36 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_-_114144599 | 0.35 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr2_-_10447771 | 0.35 |
ENST00000405333.5
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr3_+_63443076 | 0.35 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr9_+_35042213 | 0.34 |
ENST00000378745.3
ENST00000312292.6 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr3_+_42979281 | 0.33 |
ENST00000488863.5
ENST00000430121.3 |
GASK1A
|
golgi associated kinase 1A |
| chr21_+_38272410 | 0.33 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr1_-_13056575 | 0.32 |
ENST00000436041.6
|
PRAMEF27
|
PRAME family member 27 |
| chrX_-_72808210 | 0.32 |
ENST00000419795.6
ENST00000458170.1 |
FAM236D
|
family with sequence similarity 236 member D |
| chr20_-_52105644 | 0.31 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr12_-_95116967 | 0.30 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr11_-_35360050 | 0.29 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr17_-_8210203 | 0.28 |
ENST00000578549.5
ENST00000582368.5 |
AURKB
|
aurora kinase B |
| chr14_-_67412112 | 0.28 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr12_-_56741535 | 0.27 |
ENST00000647707.1
|
ENSG00000285625.1
|
novel protein |
| chr2_-_135876382 | 0.24 |
ENST00000264156.3
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr4_-_137532452 | 0.24 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr2_-_208146150 | 0.23 |
ENST00000260988.5
|
CRYGB
|
crystallin gamma B |
| chr7_-_24757926 | 0.23 |
ENST00000342947.9
ENST00000419307.6 |
GSDME
|
gasdermin E |
| chr2_-_165203870 | 0.22 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr7_-_111392915 | 0.22 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr13_-_46142834 | 0.21 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr11_+_35186820 | 0.20 |
ENST00000531110.6
ENST00000525685.6 |
CD44
|
CD44 molecule (Indian blood group) |
| chr3_+_169911566 | 0.20 |
ENST00000428432.6
ENST00000335556.7 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr9_-_21368962 | 0.19 |
ENST00000610660.1
|
IFNA13
|
interferon alpha 13 |
| chr8_-_124565699 | 0.19 |
ENST00000519168.5
|
MTSS1
|
MTSS I-BAR domain containing 1 |
| chrX_-_15314543 | 0.18 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr6_+_151239951 | 0.18 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr12_+_69825221 | 0.18 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr12_+_26011713 | 0.18 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr10_-_29634964 | 0.18 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr13_+_30422487 | 0.17 |
ENST00000638137.1
ENST00000635918.2 |
UBE2L5
|
ubiquitin conjugating enzyme E2 L5 |
| chrY_-_23694579 | 0.17 |
ENST00000343584.10
|
PRYP3
|
PTPN13 like Y-linked pseudogene 3 |
| chr12_+_69825273 | 0.17 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr11_+_5389377 | 0.16 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr3_-_186544377 | 0.16 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S |
| chr20_+_31514410 | 0.15 |
ENST00000335574.10
ENST00000340852.9 ENST00000398174.9 ENST00000466766.2 ENST00000498035.5 ENST00000344042.5 |
HM13
|
histocompatibility minor 13 |
| chr12_+_4720395 | 0.15 |
ENST00000252318.7
|
GALNT8
|
polypeptide N-acetylgalactosaminyltransferase 8 |
| chrX_-_101617921 | 0.15 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr19_-_3557563 | 0.15 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr7_-_116030735 | 0.15 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chrX_+_135309480 | 0.14 |
ENST00000635820.1
|
ETDC
|
embryonic testis differentiation homolog C |
| chr21_-_30487436 | 0.12 |
ENST00000334055.5
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr4_+_94207845 | 0.12 |
ENST00000457823.6
ENST00000354268.9 |
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_127781815 | 0.12 |
ENST00000508776.5
|
HSPA4L
|
heat shock protein family A (Hsp70) member 4 like |
| chr4_-_121164314 | 0.12 |
ENST00000057513.8
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr10_-_93482326 | 0.11 |
ENST00000359263.9
|
MYOF
|
myoferlin |
| chr10_-_93482194 | 0.11 |
ENST00000358334.9
ENST00000371488.3 |
MYOF
|
myoferlin |
| chr18_-_3845292 | 0.11 |
ENST00000400145.6
|
DLGAP1
|
DLG associated protein 1 |
| chr21_-_30492008 | 0.11 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chrX_-_49073989 | 0.11 |
ENST00000376386.3
ENST00000553851.3 |
PRAF2
|
PRA1 domain family member 2 |
| chr7_-_116030750 | 0.11 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr17_-_7404039 | 0.11 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr4_+_70242583 | 0.10 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr10_-_49188380 | 0.10 |
ENST00000374153.7
ENST00000374148.1 ENST00000374151.7 |
TMEM273
|
transmembrane protein 273 |
| chr7_+_16661182 | 0.10 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr7_-_45111673 | 0.09 |
ENST00000461363.1
ENST00000258770.8 ENST00000495078.1 ENST00000494076.5 ENST00000478532.5 ENST00000361278.7 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr4_-_103019634 | 0.09 |
ENST00000510559.1
ENST00000296422.12 ENST00000394789.7 |
SLC9B1
|
solute carrier family 9 member B1 |
| chr5_+_132873660 | 0.08 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chrX_-_31266857 | 0.08 |
ENST00000378702.8
ENST00000361471.8 |
DMD
|
dystrophin |
| chr2_-_199955464 | 0.08 |
ENST00000354611.9
|
TYW5
|
tRNA-yW synthesizing protein 5 |
| chr22_-_28711931 | 0.08 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr4_-_89835617 | 0.08 |
ENST00000611107.1
ENST00000345009.8 ENST00000505199.5 ENST00000502987.5 |
SNCA
|
synuclein alpha |
| chr2_-_169031317 | 0.08 |
ENST00000650372.1
|
ABCB11
|
ATP binding cassette subfamily B member 11 |
| chr2_+_231056845 | 0.08 |
ENST00000677230.1
ENST00000677259.1 ENST00000677180.1 ENST00000409643.6 ENST00000619128.5 ENST00000678679.1 ENST00000676740.1 ENST00000308696.11 ENST00000440838.5 ENST00000373635.9 |
PSMD1
|
proteasome 26S subunit, non-ATPase 1 |
| chr14_+_34993240 | 0.08 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr12_+_64404338 | 0.08 |
ENST00000332707.10
|
XPOT
|
exportin for tRNA |
| chr15_+_21579912 | 0.07 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr19_+_4402615 | 0.07 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr1_-_225428813 | 0.07 |
ENST00000338179.6
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr12_-_110499546 | 0.07 |
ENST00000552130.6
|
VPS29
|
VPS29 retromer complex component |
| chr10_-_5003850 | 0.07 |
ENST00000421196.7
ENST00000455190.2 ENST00000380753.8 |
AKR1C2
|
aldo-keto reductase family 1 member C2 |
| chr19_+_14583076 | 0.07 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr3_-_191282383 | 0.06 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr4_+_25160631 | 0.06 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chr9_-_92404559 | 0.06 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chrY_+_18546691 | 0.06 |
ENST00000309834.8
ENST00000307393.3 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor Y-linked 1 |
| chr17_+_4807119 | 0.06 |
ENST00000263088.11
ENST00000572940.5 |
PLD2
|
phospholipase D2 |
| chr1_-_17439657 | 0.06 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr10_+_94762673 | 0.05 |
ENST00000480405.2
ENST00000371321.9 |
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19 |
| chr19_+_3185911 | 0.05 |
ENST00000246117.9
ENST00000588428.5 |
NCLN
|
nicalin |
| chr5_+_111092172 | 0.05 |
ENST00000612402.4
|
WDR36
|
WD repeat domain 36 |
| chr19_-_45730861 | 0.05 |
ENST00000317683.4
|
FBXO46
|
F-box protein 46 |
| chrM_+_9207 | 0.05 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_-_64390852 | 0.05 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr3_-_165078480 | 0.04 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr4_+_168497044 | 0.04 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_-_87713287 | 0.04 |
ENST00000416177.1
ENST00000265724.8 ENST00000543898.5 |
ABCB1
|
ATP binding cassette subfamily B member 1 |
| chr5_-_131165272 | 0.04 |
ENST00000675491.1
ENST00000506908.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr3_+_40477107 | 0.04 |
ENST00000314686.9
ENST00000447116.6 ENST00000429348.6 ENST00000432264.4 ENST00000456778.5 |
ZNF619
|
zinc finger protein 619 |
| chr4_+_168497066 | 0.04 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr13_+_48976597 | 0.03 |
ENST00000541916.5
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr2_+_233671879 | 0.03 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr11_+_28108248 | 0.03 |
ENST00000406787.7
ENST00000403099.5 ENST00000407364.8 |
METTL15
|
methyltransferase like 15 |
| chr3_-_169769542 | 0.03 |
ENST00000330368.3
|
ACTRT3
|
actin related protein T3 |
| chr11_-_93197932 | 0.03 |
ENST00000326402.9
|
SLC36A4
|
solute carrier family 36 member 4 |
| chr8_-_63026179 | 0.03 |
ENST00000677919.1
|
GGH
|
gamma-glutamyl hydrolase |
| chr8_-_115492221 | 0.03 |
ENST00000518018.1
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr19_+_42313298 | 0.03 |
ENST00000301204.8
ENST00000673205.1 |
TMEM145
|
transmembrane protein 145 |
| chr8_+_69492793 | 0.03 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr17_+_10697576 | 0.02 |
ENST00000379774.5
|
ADPRM
|
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
| chr19_-_51417700 | 0.02 |
ENST00000529627.1
ENST00000439889.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr9_+_108862255 | 0.02 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A |
| chr11_-_95924067 | 0.02 |
ENST00000676027.1
ENST00000675489.1 ENST00000409459.5 ENST00000676261.1 ENST00000352297.11 ENST00000346299.10 ENST00000676272.1 ENST00000393223.8 ENST00000675022.1 ENST00000675362.1 ENST00000675174.1 ENST00000674989.1 ENST00000675848.1 ENST00000675652.1 ENST00000481642.6 |
MTMR2
|
myotubularin related protein 2 |
| chr19_-_51417791 | 0.02 |
ENST00000353836.9
|
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr5_-_131165231 | 0.02 |
ENST00000675100.1
ENST00000304043.10 ENST00000513012.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr10_-_126670686 | 0.02 |
ENST00000488181.3
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr1_-_178871060 | 0.02 |
ENST00000234816.7
|
ANGPTL1
|
angiopoietin like 1 |
| chr3_+_134795277 | 0.02 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1 |
| chr9_+_69145463 | 0.01 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2 |
| chr3_+_160225409 | 0.01 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chrX_+_103585478 | 0.01 |
ENST00000468024.5
ENST00000415568.2 ENST00000472484.6 ENST00000613326.4 ENST00000490644.5 ENST00000459722.1 ENST00000472745.1 ENST00000494801.5 ENST00000434216.2 |
TCEAL4
|
transcription elongation factor A like 4 |
| chr11_-_95923763 | 0.01 |
ENST00000497683.6
ENST00000470293.6 ENST00000674968.1 ENST00000484818.6 ENST00000675454.1 ENST00000675981.1 ENST00000674924.1 ENST00000676166.1 ENST00000676440.1 ENST00000675807.1 ENST00000495134.6 ENST00000675196.1 |
MTMR2
|
myotubularin related protein 2 |
| chr5_+_154755272 | 0.01 |
ENST00000518297.6
|
LARP1
|
La ribonucleoprotein 1, translational regulator |
| chr16_+_28553908 | 0.01 |
ENST00000317058.8
|
SGF29
|
SAGA complex associated factor 29 |
| chr7_-_108240049 | 0.01 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chrX_+_83861126 | 0.00 |
ENST00000621735.4
ENST00000329312.5 |
CYLC1
|
cylicin 1 |
| chr1_-_21050952 | 0.00 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 1.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.8 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 1.0 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.6 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.4 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.7 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 2.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.5 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.4 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.0 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |