Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for JDP2
Z-value: 0.54
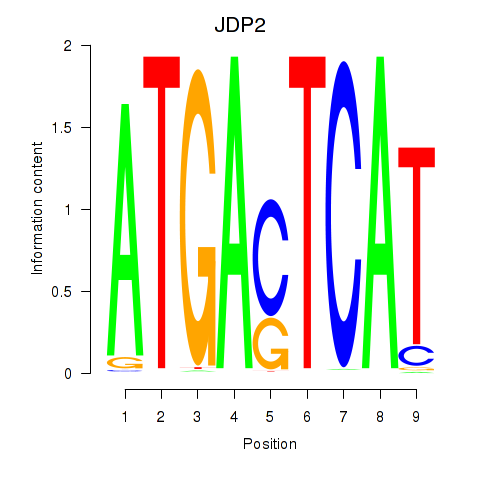
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.13 | JDP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JDP2 | hg38_v1_chr14_+_75428011_75428046 | 0.16 | 4.1e-01 | Click! |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_12904860 | 2.55 |
ENST00000336079.8
|
DDX3Y
|
DEAD-box helicase 3 Y-linked |
| chr11_-_102798148 | 2.06 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1 |
| chr9_-_122228845 | 1.15 |
ENST00000394319.8
ENST00000340587.7 |
LHX6
|
LIM homeobox 6 |
| chr6_+_130366281 | 1.04 |
ENST00000617887.4
|
TMEM200A
|
transmembrane protein 200A |
| chr11_-_102955705 | 0.76 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr10_+_17228215 | 0.73 |
ENST00000544301.7
|
VIM
|
vimentin |
| chr15_+_88638947 | 0.72 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr15_+_88639009 | 0.68 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr12_-_119804472 | 0.66 |
ENST00000678087.1
ENST00000677993.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr12_-_119804298 | 0.65 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr16_+_15502266 | 0.65 |
ENST00000452191.6
|
BMERB1
|
bMERB domain containing 1 |
| chr6_+_33075952 | 0.63 |
ENST00000418931.7
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr1_-_204152010 | 0.63 |
ENST00000367202.9
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_204151884 | 0.63 |
ENST00000367201.7
|
ETNK2
|
ethanolamine kinase 2 |
| chr8_-_27614681 | 0.59 |
ENST00000519472.5
ENST00000523589.5 ENST00000522413.5 ENST00000523396.1 ENST00000316403.15 |
CLU
|
clusterin |
| chr19_-_35528221 | 0.51 |
ENST00000588674.5
ENST00000452271.7 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr9_-_120876356 | 0.51 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr17_+_4950147 | 0.51 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr3_-_18425295 | 0.48 |
ENST00000338745.11
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr9_-_120877026 | 0.48 |
ENST00000436309.5
|
PHF19
|
PHD finger protein 19 |
| chr10_-_25062279 | 0.47 |
ENST00000615958.4
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr10_+_125973373 | 0.45 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr19_-_35501878 | 0.44 |
ENST00000593342.5
ENST00000601650.1 ENST00000408915.6 |
DMKN
|
dermokine |
| chr2_+_10122315 | 0.40 |
ENST00000360566.6
|
RRM2
|
ribonucleotide reductase regulatory subunit M2 |
| chr6_-_118710065 | 0.38 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr7_-_151024423 | 0.36 |
ENST00000469530.4
ENST00000639579.1 |
ATG9B
|
autophagy related 9B |
| chr18_-_76495191 | 0.34 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr3_+_98353854 | 0.34 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor family 5 subfamily K member 4 |
| chr9_-_113303271 | 0.33 |
ENST00000297894.5
ENST00000489339.2 |
RNF183
|
ring finger protein 183 |
| chr6_-_161274042 | 0.33 |
ENST00000320285.9
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr6_-_161274010 | 0.33 |
ENST00000366911.9
ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_-_43781249 | 0.33 |
ENST00000615047.4
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr11_-_82997477 | 0.32 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr20_-_17558811 | 0.31 |
ENST00000536626.7
ENST00000377868.6 |
BFSP1
|
beaded filament structural protein 1 |
| chr12_-_54419259 | 0.29 |
ENST00000293379.9
|
ITGA5
|
integrin subunit alpha 5 |
| chr4_-_99435134 | 0.28 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_-_52949818 | 0.26 |
ENST00000546897.5
ENST00000552551.5 |
KRT8
|
keratin 8 |
| chr3_-_48595267 | 0.25 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr1_-_246417404 | 0.24 |
ENST00000630181.2
|
SMYD3
|
SET and MYND domain containing 3 |
| chr4_-_121164314 | 0.23 |
ENST00000057513.8
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr16_+_50266530 | 0.23 |
ENST00000566433.6
ENST00000394697.7 ENST00000673801.1 |
ADCY7
|
adenylate cyclase 7 |
| chr11_-_83071819 | 0.23 |
ENST00000524635.1
ENST00000526205.5 ENST00000533486.5 ENST00000533276.6 ENST00000527633.6 |
RAB30
|
RAB30, member RAS oncogene family |
| chr7_+_97732046 | 0.23 |
ENST00000350485.8
ENST00000346867.4 ENST00000319273.10 |
TAC1
|
tachykinin precursor 1 |
| chr4_-_993430 | 0.22 |
ENST00000361661.6
ENST00000622731.4 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr3_-_196270540 | 0.22 |
ENST00000419333.5
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr7_+_28685968 | 0.22 |
ENST00000396298.6
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr17_-_28718147 | 0.22 |
ENST00000436730.7
ENST00000625712.2 ENST00000450529.5 ENST00000583538.5 ENST00000419712.7 ENST00000580843.6 ENST00000582934.1 ENST00000415040.6 ENST00000353676.9 ENST00000453384.7 |
RAB34
|
RAB34, member RAS oncogene family |
| chr3_-_18424533 | 0.21 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr17_-_28718405 | 0.21 |
ENST00000430132.6
ENST00000301043.10 ENST00000412625.5 |
RAB34
|
RAB34, member RAS oncogene family |
| chr18_+_24113341 | 0.21 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr4_-_99435396 | 0.20 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr19_-_45424364 | 0.20 |
ENST00000589165.5
|
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr16_+_31355165 | 0.20 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr8_+_69492793 | 0.20 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr6_+_71886703 | 0.19 |
ENST00000491071.6
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr1_+_213051229 | 0.19 |
ENST00000615329.4
ENST00000543354.5 ENST00000614059.4 ENST00000543470.5 ENST00000366960.8 ENST00000366959.4 |
RPS6KC1
|
ribosomal protein S6 kinase C1 |
| chr8_+_15540223 | 0.18 |
ENST00000382020.8
ENST00000506802.5 ENST00000503731.6 ENST00000509380.5 |
TUSC3
|
tumor suppressor candidate 3 |
| chr4_-_99435336 | 0.18 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_-_28717860 | 0.18 |
ENST00000395245.9
|
RAB34
|
RAB34, member RAS oncogene family |
| chr6_+_292050 | 0.18 |
ENST00000344450.9
|
DUSP22
|
dual specificity phosphatase 22 |
| chr5_-_76623391 | 0.18 |
ENST00000296641.5
ENST00000504899.1 |
F2RL2
|
coagulation factor II thrombin receptor like 2 |
| chrX_-_15270031 | 0.17 |
ENST00000380483.7
ENST00000380485.7 ENST00000380488.9 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr3_-_45915698 | 0.17 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr2_+_28392802 | 0.16 |
ENST00000379619.5
ENST00000264716.9 |
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr3_+_191329342 | 0.16 |
ENST00000392455.9
|
CCDC50
|
coiled-coil domain containing 50 |
| chr11_-_69819410 | 0.15 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr17_+_68249200 | 0.15 |
ENST00000577985.5
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr7_+_832470 | 0.14 |
ENST00000401592.6
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chrX_+_134237047 | 0.14 |
ENST00000370809.4
ENST00000517294.5 |
CCDC160
|
coiled-coil domain containing 160 |
| chr18_+_63775369 | 0.14 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr2_+_47369301 | 0.14 |
ENST00000263735.9
|
EPCAM
|
epithelial cell adhesion molecule |
| chr15_-_25865076 | 0.14 |
ENST00000619904.1
|
ATP10A
|
ATPase phospholipid transporting 10A (putative) |
| chr7_-_80922354 | 0.14 |
ENST00000419255.6
|
SEMA3C
|
semaphorin 3C |
| chr8_-_124372686 | 0.13 |
ENST00000297632.8
|
TMEM65
|
transmembrane protein 65 |
| chr17_+_77450737 | 0.13 |
ENST00000541152.6
ENST00000591704.5 |
SEPTIN9
|
septin 9 |
| chr12_-_130716264 | 0.13 |
ENST00000643940.1
|
RIMBP2
|
RIMS binding protein 2 |
| chr17_-_28717792 | 0.13 |
ENST00000636772.1
|
RAB34
|
RAB34, member RAS oncogene family |
| chr6_-_149484965 | 0.13 |
ENST00000409806.8
|
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr11_-_57649932 | 0.13 |
ENST00000524669.5
|
YPEL4
|
yippee like 4 |
| chr18_+_63775395 | 0.13 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr2_-_219308963 | 0.13 |
ENST00000423636.6
ENST00000442029.5 ENST00000412847.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr5_+_35856883 | 0.13 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr1_-_153549120 | 0.12 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_+_832488 | 0.12 |
ENST00000405266.5
ENST00000403868.5 ENST00000425407.6 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr15_+_86079863 | 0.12 |
ENST00000614907.3
ENST00000441037.7 |
AGBL1
|
ATP/GTP binding protein like 1 |
| chr20_+_49812697 | 0.12 |
ENST00000417961.5
|
SLC9A8
|
solute carrier family 9 member A8 |
| chr9_-_72364504 | 0.12 |
ENST00000237937.7
ENST00000343431.6 ENST00000376956.3 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr10_+_119651372 | 0.12 |
ENST00000369085.8
|
BAG3
|
BAG cochaperone 3 |
| chr5_+_73813518 | 0.12 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr17_-_1649854 | 0.12 |
ENST00000301336.7
|
RILP
|
Rab interacting lysosomal protein |
| chr2_+_219627394 | 0.12 |
ENST00000373760.6
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr3_+_69084973 | 0.12 |
ENST00000478935.1
|
ARL6IP5
|
ADP ribosylation factor like GTPase 6 interacting protein 5 |
| chr9_-_34662654 | 0.12 |
ENST00000259631.5
|
CCL27
|
C-C motif chemokine ligand 27 |
| chr1_-_243843226 | 0.12 |
ENST00000336199.9
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr11_-_57649894 | 0.11 |
ENST00000534810.3
ENST00000300022.8 |
YPEL4
|
yippee like 4 |
| chr4_+_73740541 | 0.11 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr8_-_70071226 | 0.11 |
ENST00000276594.3
|
PRDM14
|
PR/SET domain 14 |
| chr8_-_140764386 | 0.11 |
ENST00000520151.5
ENST00000519024.5 ENST00000519465.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_219627622 | 0.11 |
ENST00000358055.8
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr1_-_161132577 | 0.11 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr16_-_28506826 | 0.11 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr2_+_219627650 | 0.11 |
ENST00000317151.7
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr6_+_10521337 | 0.11 |
ENST00000495262.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr20_+_49812818 | 0.11 |
ENST00000361573.3
|
SLC9A8
|
solute carrier family 9 member A8 |
| chr1_+_183186238 | 0.11 |
ENST00000493293.5
ENST00000264144.5 |
LAMC2
|
laminin subunit gamma 2 |
| chr3_-_98522514 | 0.11 |
ENST00000503004.5
ENST00000506575.1 ENST00000513452.5 ENST00000515620.5 |
CLDND1
|
claudin domain containing 1 |
| chr6_-_83431038 | 0.10 |
ENST00000369705.4
|
ME1
|
malic enzyme 1 |
| chr2_+_219627565 | 0.10 |
ENST00000273063.10
|
SLC4A3
|
solute carrier family 4 member 3 |
| chr1_+_222815021 | 0.10 |
ENST00000675850.1
ENST00000495684.2 |
DISP1
|
dispatched RND transporter family member 1 |
| chr1_-_153549238 | 0.10 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chr20_+_32358303 | 0.10 |
ENST00000651418.1
ENST00000375687.10 ENST00000542461.5 ENST00000613218.4 ENST00000646367.1 ENST00000620121.4 |
ASXL1
|
ASXL transcriptional regulator 1 |
| chr5_+_136058849 | 0.10 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr1_+_222815065 | 0.10 |
ENST00000675039.1
ENST00000675961.1 |
DISP1
|
dispatched RND transporter family member 1 |
| chr1_-_209651291 | 0.09 |
ENST00000391911.5
ENST00000415782.1 |
LAMB3
|
laminin subunit beta 3 |
| chr3_-_98522869 | 0.09 |
ENST00000502288.5
ENST00000512147.5 ENST00000341181.11 ENST00000510541.5 ENST00000503621.5 ENST00000511081.5 ENST00000507874.5 ENST00000502299.5 ENST00000508659.5 ENST00000510545.5 ENST00000511667.5 ENST00000394185.6 |
CLDND1
|
claudin domain containing 1 |
| chr17_-_41140487 | 0.09 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr7_+_120950763 | 0.09 |
ENST00000339121.9
ENST00000315870.10 ENST00000445699.5 |
ING3
|
inhibitor of growth family member 3 |
| chr14_-_73760259 | 0.09 |
ENST00000286523.9
ENST00000435371.1 |
MIDEAS
|
mitotic deacetylase associated SANT domain protein |
| chr17_-_75270409 | 0.09 |
ENST00000618645.5
|
MIF4GD
|
MIF4G domain containing |
| chr1_+_15617415 | 0.09 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2 |
| chr3_+_57060658 | 0.08 |
ENST00000334325.2
|
SPATA12
|
spermatogenesis associated 12 |
| chr4_+_75556048 | 0.08 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr10_+_113709261 | 0.08 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chr2_+_74529923 | 0.08 |
ENST00000258080.8
ENST00000352222.7 |
HTRA2
|
HtrA serine peptidase 2 |
| chr16_-_31150058 | 0.08 |
ENST00000569305.1
ENST00000268281.9 ENST00000418068.6 |
PRSS36
|
serine protease 36 |
| chr11_-_84317296 | 0.08 |
ENST00000280241.12
ENST00000398301.6 |
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr3_-_151316795 | 0.08 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr17_+_76385256 | 0.08 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr15_-_22980334 | 0.08 |
ENST00000610365.4
ENST00000617928.5 ENST00000611832.4 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chr4_+_174918355 | 0.07 |
ENST00000505141.5
ENST00000359240.7 ENST00000615367.4 ENST00000445694.5 ENST00000618444.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr13_-_41019289 | 0.07 |
ENST00000239882.7
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr4_-_145098541 | 0.07 |
ENST00000613466.4
ENST00000514390.5 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr3_+_191329020 | 0.07 |
ENST00000392456.4
|
CCDC50
|
coiled-coil domain containing 50 |
| chr5_+_145937793 | 0.07 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr2_-_152098810 | 0.07 |
ENST00000636442.1
ENST00000638005.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr16_+_30065753 | 0.07 |
ENST00000642816.3
ENST00000643777.4 ENST00000569798.5 |
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr8_-_109975757 | 0.07 |
ENST00000524391.6
|
KCNV1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr2_+_74530018 | 0.07 |
ENST00000437202.1
|
HTRA2
|
HtrA serine peptidase 2 |
| chr19_+_13731744 | 0.06 |
ENST00000586600.5
|
CCDC130
|
coiled-coil domain containing 130 |
| chr11_-_102874974 | 0.06 |
ENST00000571244.3
|
MMP12
|
matrix metallopeptidase 12 |
| chr13_+_53028806 | 0.06 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr1_-_161132659 | 0.06 |
ENST00000368006.8
ENST00000490843.6 ENST00000545495.5 |
DEDD
|
death effector domain containing |
| chr17_+_77319465 | 0.06 |
ENST00000329047.13
|
SEPTIN9
|
septin 9 |
| chrX_+_47193796 | 0.06 |
ENST00000442035.5
ENST00000335972.11 ENST00000457753.5 |
UBA1
|
ubiquitin like modifier activating enzyme 1 |
| chrX_+_57592011 | 0.06 |
ENST00000374888.3
|
ZXDB
|
zinc finger X-linked duplicated B |
| chr4_+_174918400 | 0.06 |
ENST00000404450.8
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr11_-_83071917 | 0.05 |
ENST00000534141.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr2_-_65366650 | 0.05 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr3_-_9878488 | 0.05 |
ENST00000443115.1
|
CIDEC
|
cell death inducing DFFA like effector c |
| chr11_-_102843597 | 0.05 |
ENST00000299855.10
|
MMP3
|
matrix metallopeptidase 3 |
| chr6_-_56954747 | 0.05 |
ENST00000680361.1
|
DST
|
dystonin |
| chr9_-_89178810 | 0.05 |
ENST00000375835.9
|
SHC3
|
SHC adaptor protein 3 |
| chr16_+_30065777 | 0.05 |
ENST00000395240.7
ENST00000566846.5 |
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr19_-_51034892 | 0.05 |
ENST00000319590.8
ENST00000250351.4 |
KLK12
|
kallikrein related peptidase 12 |
| chr3_+_48466222 | 0.04 |
ENST00000625293.3
ENST00000492235.2 ENST00000635452.2 ENST00000456089.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr2_+_46698909 | 0.04 |
ENST00000650611.1
ENST00000306503.5 |
LINC01118
SOCS5
|
long intergenic non-protein coding RNA 1118 suppressor of cytokine signaling 5 |
| chr4_-_151226427 | 0.04 |
ENST00000304527.8
ENST00000409598.8 |
SH3D19
|
SH3 domain containing 19 |
| chr19_-_45782388 | 0.04 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr8_-_42768781 | 0.04 |
ENST00000276410.7
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr16_+_31355215 | 0.04 |
ENST00000562522.2
|
ITGAX
|
integrin subunit alpha X |
| chr11_-_2903490 | 0.04 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chr17_-_29930062 | 0.04 |
ENST00000579954.1
ENST00000269033.7 ENST00000540801.6 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr8_-_42768602 | 0.04 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr15_+_75043263 | 0.04 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr16_+_14186707 | 0.03 |
ENST00000572567.5
|
MRTFB
|
myocardin related transcription factor B |
| chr1_+_156126525 | 0.03 |
ENST00000504687.6
ENST00000473598.6 |
LMNA
|
lamin A/C |
| chr1_+_89524871 | 0.03 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr3_+_69084929 | 0.03 |
ENST00000273258.4
|
ARL6IP5
|
ADP ribosylation factor like GTPase 6 interacting protein 5 |
| chrX_+_154542194 | 0.03 |
ENST00000618670.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr10_+_132064685 | 0.03 |
ENST00000670120.1
ENST00000653512.1 ENST00000659050.1 ENST00000653623.1 ENST00000664697.1 ENST00000660144.1 ENST00000670443.1 |
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr3_-_194369731 | 0.03 |
ENST00000428839.1
ENST00000347624.4 |
LRRC15
|
leucine rich repeat containing 15 |
| chr4_+_83535914 | 0.03 |
ENST00000611707.4
|
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr13_-_35855758 | 0.03 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr3_-_31981228 | 0.03 |
ENST00000396556.7
ENST00000438237.6 |
OSBPL10
|
oxysterol binding protein like 10 |
| chr4_+_83536097 | 0.03 |
ENST00000395226.6
ENST00000264409.5 |
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr1_+_151254709 | 0.03 |
ENST00000368881.8
ENST00000368884.8 |
PSMD4
|
proteasome 26S subunit, non-ATPase 4 |
| chr20_+_44401269 | 0.03 |
ENST00000443598.6
ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr10_+_132065937 | 0.03 |
ENST00000658847.1
ENST00000666974.1 |
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr19_-_51034840 | 0.03 |
ENST00000529888.5
|
KLK12
|
kallikrein related peptidase 12 |
| chr17_-_40999903 | 0.03 |
ENST00000391587.3
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr11_+_102112445 | 0.03 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator |
| chr17_+_81098118 | 0.03 |
ENST00000416299.6
|
BAIAP2
|
BAR/IMD domain containing adaptor protein 2 |
| chr10_+_132066003 | 0.03 |
ENST00000657318.1
ENST00000666210.1 |
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr16_-_53052849 | 0.03 |
ENST00000619363.2
|
ENSG00000277639.2
|
novel protein |
| chr2_-_223602284 | 0.02 |
ENST00000421386.1
ENST00000305409.3 ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr14_+_103334803 | 0.02 |
ENST00000561325.5
ENST00000392715.6 ENST00000559130.5 ENST00000559532.5 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr13_-_35855627 | 0.02 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr7_-_14903319 | 0.02 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta |
| chr10_+_133087883 | 0.02 |
ENST00000392607.8
|
ADGRA1
|
adhesion G protein-coupled receptor A1 |
| chr6_+_45328203 | 0.02 |
ENST00000371432.7
ENST00000647337.2 ENST00000371438.5 |
RUNX2
|
RUNX family transcription factor 2 |
| chr2_+_201257980 | 0.02 |
ENST00000447616.5
ENST00000358485.8 ENST00000392266.7 |
CASP8
|
caspase 8 |
| chr10_-_73625951 | 0.02 |
ENST00000433394.1
ENST00000422491.7 |
USP54
|
ubiquitin specific peptidase 54 |
| chr10_-_103479240 | 0.02 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr6_-_49787265 | 0.02 |
ENST00000304801.6
|
PGK2
|
phosphoglycerate kinase 2 |
| chr12_-_7665897 | 0.01 |
ENST00000229304.5
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr8_-_67062120 | 0.01 |
ENST00000357849.9
|
COPS5
|
COP9 signalosome subunit 5 |
| chr2_+_169069537 | 0.01 |
ENST00000428522.5
ENST00000450153.1 ENST00000674881.1 ENST00000421653.5 |
DHRS9
|
dehydrogenase/reductase 9 |
| chr11_+_67483019 | 0.01 |
ENST00000279146.8
ENST00000528641.7 ENST00000682324.1 ENST00000684006.1 ENST00000683237.1 ENST00000684657.1 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr5_+_146203593 | 0.01 |
ENST00000265271.7
|
RBM27
|
RNA binding motif protein 27 |
| chr12_-_102197827 | 0.01 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 0.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.6 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.2 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 1.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 2.1 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0030718 | inner cell mass cell fate commitment(GO:0001827) germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.8 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 2.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.3 | GO:0090292 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 1.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:0033631 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.6 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 1.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 0.7 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 2.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |